The Glycosyltransferases of LPS Core: A Review of Four Heptosyltransferase Enzymes in Context
Abstract
:1. Introduction
2. Glycosyltransferases
2.1. Glycosyltransferase Structural Folds
2.1.1. GT-A Structural Fold
2.1.2. GT-B Structural Fold
2.1.3. GT-C Structural Fold
2.1.4. Catalytic Mechanisms
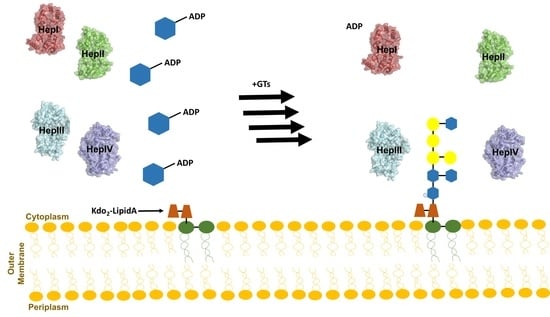
3. Core Heptosyltransferase Enzymes
3.1. Multiple Sequence Alignment (MSA) of Heptosyltransferase Enzymes
3.2. Heptosyltransferase I
3.2.1. Crystal Structures of HepI
3.2.2. Inhibition of HepI
3.2.3. Investigations of HepI Protein Dynamics
4. Conclusions
Author Contributions
Conflicts of Interest
References
- Barbier, F.; Luyt, C. Understanding resistance. Intensiv. Care Med. 2016, 42, 2080–2083. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). Antibiotic Resistance Threats in the United States, 2013; Technical Report; CDC; U.S. Department of Health and Human Services: Atlanta, GA, USA, 2013. Available online: http://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf (accessed on 28 September 2017).
- World Health Organization (WHO). Antibiotic Resistance; Fact Sheet. World Health Organization (WHO), 2016. Available online: http://www.who.int/mediacentre/factsheets/antibiotic-resistance/en/ (accessed on 28 September 2017).
- Ruiz, N.; Kahne, D.; Silhavy, T.J. Advances in understanding bacterial outer-membrane biogenesis. Nat. Rev. Microbiol. 2006, 4, 57–66. [Google Scholar] [PubMed]
- Nikaido, H. Molecular Basis of Bacterial Outer Membrane Permeability Revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef] [PubMed]
- Piek, S.; Kahler, C.M. A comparison of the endotoxin biosynthesis and protein oxidation pathways in the biogenesis of the outer membrane of Escerichia coli and Neisseria meningitidis. Front. Cell. Infect. Microbiol. 2012, 2, 162. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.; Reynolds, M.C.; Trent, S.M.; Bishop, R.E. Lipid A Modification Systems in Gram-Negative Bacteria. Annu. Rev. Biochem. 2007, 76, 295–329. [Google Scholar] [CrossRef] [PubMed]
- Schnaitman, C.A.; Klena, J.D. Genetics of Lipopolysaccharide Biosynthesis in Enteric Bacteria. Microbiol. Rev. 1993, 57, 655–682. [Google Scholar] [PubMed]
- Raetz, C.R.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef] [PubMed]
- Coleman, W.G.; Leive, L. Two mutations which affect the barrier function of the Escherichia coli K-12 outer membrane. J. Bacteriol. 1979, 139, 899–910. [Google Scholar] [PubMed]
- Stenutz, R.; Weintraub, A.; Widmalm, G. The structures of Escherichia coli O-polysaccharide antigens. FEMS Microbiol. Rev. 2006, 30, 382–403. [Google Scholar] [CrossRef] [PubMed]
- Lerouge, I.; Vanderleyden, J. O-antigen structural variation: Mechanisms and possible roles in animal/plant–microbe interactions. FEMS Microbiol. Rev. 2002, 26, 17–47. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.N.; Chaudhuri, K. Lipopolysaccharides of Vibrio cholerae: I. Physical and chemical characterization. Biochim. Biophys. Acta 2003, 1639, 65–79. [Google Scholar] [CrossRef] [PubMed]
- Caroff, M.; Karibian, D. Structure of bacterial lipopolysaccharides. Carbohydr. Res. 2003, 338, 2341–2347. [Google Scholar] [CrossRef]
- Heinrichs, D.E.; Yethon, J.A.; Whitfield, C. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol. Microbiol. 1998, 30, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.; Garrett, T.A.; Reynolds, C.M.; Shaw, W.A.; Moore, J.D.; Dale, C.; Smith, J.; Ribeiro, A.A.; Murphy, R.C.; Ulevitch, R.J.; et al. Kdo2-Lipid A of Escherichia coli, a defined endotoxin that activates microphages via TLR-4. J. Lipid Res. 2006, 47, 1097–1111. [Google Scholar] [CrossRef] [PubMed]
- Whitfield, C.; Trent, M. Biosynthesis and export of bacterial lipopolysaccharides. Annu. Rev. Biochem. 2014, 83, 99–128. [Google Scholar] [CrossRef] [PubMed]
- Delcour, A.H. Outer membrane permeability and antibiotic resistance. Biochim. Biophys. Acta 2009, 1794, 808–816. [Google Scholar] [CrossRef] [PubMed]
- Klein, G.; Lindner, B.; Brabetz, W.; Brade, H.; Raina, S. Escherichia coli K-12 Suppressor-free Mutants Lacking Early Glycosyltransferase and Late Acyltransferase: Minimal Lipopolysaccharide Structure and Induction of Envelope Stress Response. J. Biol. Chem. 2009, 284, 15369–15389. [Google Scholar] [CrossRef] [PubMed]
- Kanipes, M.I.; Holder, L.C.; Corcoran, A.T.; Moran, A.P.; Guerry, P. A Deep-Rough Mutant of Campylobacter jejuni 81-176 Is Noninvasive for Intestinal Epithelial Cells. Infect. Immun. 2004, 72, 2452–2455. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R. Bacterial lipopolysaccharides: A remarkable family of bioactive macroamphiphiles. In Escherichia coli and Salmonella: Cellular and Molecular Biology; Neidhardt, F.C., Curtiss, R., Ingraham, J.L., III, Lin, E.C.C., Low, K.B., Magasanik, B., Reznikoff, W.S., Riley, M., Schaechter, M., Umbarger, H.E., Eds.; American Society for Microbiology: Washington, DC, USA, 1996. [Google Scholar]
- Coutinho, P.M.; Deleury, E.; Davies, G.J.; Henrissat, B. An Evolving Hierarchical Family Classification for Glycosyltransferases. J. Mol. Biol. 2003, 328, 307–317. [Google Scholar] [CrossRef]
- Varki, A.; Cummings, R.; Esko, J.; Freeze, H.; Hart, G.; Marth, J. (Eds.) Essentials of Glycobiology; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1999; Chapter 17 (Glycosyltransferases); pp. 253–262. [Google Scholar]
- Albesa-Jové, D.; Giganti, D.; Jackson, M.; Alzari, P.; Guerin, M. Structure-function relationships of membrane-associated GT-B glycosyltransferases. Glycobiology 2014, 24, 108–124. [Google Scholar] [CrossRef] [PubMed]
- Lairson, L.; Henrissat, B.; Davies, G.; Withers, S. Glycosyltransferases: Structures, functions, and mechanisms. Annu. Rev. Biochem. 2008, 77, 521–555. [Google Scholar] [CrossRef] [PubMed]
- Lombard, V.; Golaconda Ramulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef] [PubMed]
- Sinnott, M.L. Catalytic mechanism of enzymic glycosyl transfer. Chem. Rev. 1990, 90, 1171–1202. [Google Scholar] [CrossRef]
- Charnock, S.J.; Davies, G.J. Structure of the Nucleotide-Diphospho-Sugar Transferase, SpsA from Bacillus subtilis, in Native and Nucleotide-Complexed Forms. Biochemistry 1999, 38, 6380–6385. [Google Scholar] [CrossRef] [PubMed]
- Pak, J.E.; Arnoux, P.; Zhou, S.; Sivarajah, P.; Satkunarajah, M.; Xing, X.; Rini, J.M. X-ray Crystal Structure of Leukocyte Type Core 2 β1,6-N-Acetylglucosaminyltransferase: Evidencec for a Convergence of Metal Ion-independent Glycosyltransferase Mechanism. J. Biol. Chem. 2006, 281, 26693–26701. [Google Scholar] [CrossRef] [PubMed]
- Urresti, S.; Albesa-Jové, D.; Schaeffer, F.; Pham, H.T.; Kaur, D.; Gest, P.; van der Woerd, M.J.; Carreras-González, A.; López-Fernández, S.; Alzari, P.M.; et al. Mechanistic Insights into the Retaining Glucosyl-3-phosphoglycerate Synthase from Mycobacteria. J. Biol. Chem. 2012, 287, 24649–24661. [Google Scholar] [CrossRef] [PubMed]
- Grizot, S.; Salem, M.; Vongsouthi, V.; Durand, L.; Moreau, F.; Dohi, H.; Vincent, S.; Escaich, S.; Ducruix, A. Structure of the Escherichia coli Heptosyltransferase WaaC: Binary Complexes with ADP and ADP-2-deoxy-2-Fluoro Heptose. J. Mol. Biol. 2006, 363, 383–394. [Google Scholar] [CrossRef] [PubMed]
- Vrielink, A.; Rüger, W.; Driessen, H.P.; Freemont, P.S. Crystal structure of the DNA modifying enzyme beta-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose. EMBO J. 1994, 13, 3413–3422. [Google Scholar] [PubMed]
- Vetting, M.W.; Frantom, P.A.; Blanchard, J.S. Structural and Enzymatic Analysis of MshA from Corynebacterium glutamicum: Substrate-assisted Cataylsis. J. Biol. Chem. 2008, 283, 15834–15844. [Google Scholar] [CrossRef] [PubMed]
- Mulichak, A.M.; Losey, H.C.; Lu, W.; Wawrzak, Z.; Walsh, C.T.; Garavito, R.M. Structure of the TDP-epi-vancosaminyltransferase GtfA from the chloroeremomycin biosynthetic pathway. Proc. Natl. Acad. Sci. USA 2003, 100, 9238–9243. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Mushegian, A. Three monophyletic superfamilies account for the majority of the known glycosyltransferases. Protein Sci. 2003, 12, 1418–1431. [Google Scholar] [CrossRef] [PubMed]
- Lizak, C.; Gerber, S.; Numao, S.; Aebi, M.; Locher, K.P. X-ray structure of a bacterial oligosaccharyltransferase. Nature 2011, 474, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Kozmon, S.; Tvaroška, I. Catalytic Mechanism of Glycosyltranferases: Hybrid Quantum Mechanical/Molecular Mechanical Study of the Inverting N-Acetylglucosaminyltransferase I. J. Am. Chem. Soc. 2006, 128, 16921–16927. [Google Scholar] [CrossRef] [PubMed]
- Taylor Ringia, E.A.; Schramm, V.L. Transition states and inhibitors of the purine nucleoside phosphorylase family. Curr. Top. Med. Chem. 2005, 5, 1237–1258. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Cervellera, V.; Ardèvol, A.; Boero, M.; Planas, A.; Rovira, C. Formation of a Covalent Glycosyl–Enzyme Species in a Retaining Glycosyltransferase. Chem. Eur. J. 2013, 19, 14018–14023. [Google Scholar] [CrossRef] [PubMed]
- Ardèvol, A.; Iglesias-Fernández, J.; Rojas-Cervellera, V.; Rovira, C. The reaction mechanism of retaining glycosyltransferases. Biochem. Soc. Trans. 2016, 44, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Soya, N.; Fang, Y.; Palcic, M.M.; Klassen, J.S. Trapping and characterization of covalent intermediates of mutant retaining glycosyltransferases. Glycobiology 2011, 21, 547–552. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.S.; Hong, S.Y.; Errey, J.C.; Izumi, A.; Davies, G.J.; Davis, B.G. Mechanistic evidence for a front-side, SNi-type reaction in a retaining glycosyltransferase. Nat. Chem. Biol. 2011, 7, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Frantom, P.A.; Coward, J.K.; Blanchard, J.S. UDP-(5F)-GlcNAc Acts as a Slow-Binding Inhibitor of MshA, a Retaining Glycosyltransferase. J. Am. Chem. Soc. 2010, 132, 6626–6627. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Quinn, P.J. Lipopolysaccharide: Biosynthetic pathway and structure modification. Prog. Lipid Res. 2010, 49, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Schrinner, E. Surface Structures of Microorganisms and Their Interactions with the Mammalian Host. In Proceedings of the Eighteenth Workshop Conference, Hoechst, Schloss Ringberg, Germany, 20–23 October 1987; John Wiley & Sons: Hoboken, NJ, USA, 1988; Volume 18. [Google Scholar]
- Zhao, X.; Wenzel, C.Q.; Lam, J.S. Nonradiolabeling Assay for WaaP, an Essential Sugar Kinase Involved in Biosynthesis of Core Lipopolysaccharide of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2002, 46, 2035–2037. [Google Scholar] [CrossRef] [PubMed]
- Knirel, Y.; Anisimov, A. Lipopolysaccharide of Yersinia pestis, the Cause of Plague: Structure, Genetics, Biological Properties. Acta Nat. 2012, 4, 46–58. [Google Scholar]
- Reeves, P.P.; Wang, L. Genomic organization of LPS-specific loci. Curr. Top. Microbiol. Immunol. 2002, 264, 109–135. [Google Scholar]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.; Blackshields, G.; Brown, N.; Chenna, R.; McGettigan, P.; McWilliam, H.; Valentin, F.; Wallace, I.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Mudapaka, J.; Taylor, E.A. Cloning and Characterization of the Escherichia coli Heptosyltransferase III: Exploring Substrate Specificity in Lipopolysaccharide Core Biosynthesis. FEBS Lett. 2015, 589, 1423–1429. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef] [PubMed]
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 2010, 5, 725–738. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [PubMed]
- Kadrmas, J.L.; Raetz, C.R.H. Enzymatic Synthesis of Lipopolysaccharide in Escherichia coli: Purification and Properties of Heptosyltransferase I. J. Biol. Chem. 1998, 273, 2799–2807. [Google Scholar] [CrossRef] [PubMed]
- Czyzyk, D.J.; Liu, C.; Taylor, E.A. Lipopolysaccharide Biosynthesis without the Lipids: Recognition Promiscuity of Escherichia coli Heptosyltransferase I. Biochemistry 2011, 50, 10570–10572. [Google Scholar] [CrossRef] [PubMed]
- Larivière, L.; Gueguen-Chaignon, V.; Moréra, S. Crystal Structures of the T4 Phage β-Glucosyltransferase and the D100A Mutant in Complex with UDP-glucose: Glucose Binding and Identification of the Catalytic Base for a Direct Displacement Mechanism. J. Mol. Biol. 2003, 330, 1077–1086. [Google Scholar] [CrossRef]
- Moreau, F.; Desroy, N.; Genevard, J.M.; Vongsouthi, V.; Gerusz, V.; Le Fralliec, G.; Oliveira, C.; Floquet, S.; Denis, A.; Escaich, S.; et al. Discovery of New Gram-negative Antivirulence Drugs: Structure and Properties of Novel E. coli WaaC Inhibitors. Bioorg. Med. Chem. Lett. 2008, 18, 4022–4026. [Google Scholar] [CrossRef] [PubMed]
- Durka, M.; Buffet, K.; Iehl, J.; Holler, M.; Nierengarten, J.F.; Vincent, S.P. The Inhibition of Liposaccharide Heptosyltransferase WaaC with Multivalent Glycosylated Fullerenes: A New Mode of Glycosyltransferase Inhibition. Chem. Eur. J. 2012, 18, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Tikad, A.; Fu, H.; Sevrain, C.M.; Laurent, S.; Nierengarten, J.; Vincent, S.P. Mechanistic Insight into Heptosyltransferase Inhibition by using Kdo Multivalent Glycoclusters. Chem. Eur. J. 2016, 22, 13147–13155. [Google Scholar] [CrossRef] [PubMed]
- Czyzyk, D.J.; Sawant, S.; Ramirez-Mondragon, C.A.; Hingorani, M.M.; Taylor, E.A. Escherichia coli Heptosyltransferase I: Investigation of Protein Dynamics of a GT-B Structural Enzyme. Biochemistry 2013, 52, 5158–5160. [Google Scholar]
- Garrity, J.D.; Pauff, J.M.; Crowder, M.W. Probing the Dynamics of a Mobile Loop above the Active Site of L1, a Metallo-β-lactamase from Stenotrophomonas maltophilia, via Site-directed Mutagenesis and Stopped-flow Fluorescence Spectroscopy. J. Biol. Chem. 2004, 279, 39663–39670. [Google Scholar] [CrossRef] [PubMed]
- Dumitraşcu, L.; Stănciuc, N.; Bahrim, G.E.; Ciumac, A.; Aprodu, I. pH and heat-dependent behaviour of glucose oxidase down to single molecule level by combined fluorescence spectroscopy and molecular modelling. J. Sci. Food Agric. 2016, 96, 1906–1914. [Google Scholar] [CrossRef] [PubMed]
- Hogue, C.W.V.; Doublié, S.; Xue, H.; Wong, J.T.; Carter, C.W., Jr.; Szabo, A.G. A Concerted Tryptophanyl-adenylate-dependent Conformational Change in Bacillus subtilis Tryptophanyl-tRNA Synthetase Revealed by the Fluorescence of Trp92. J. Mol. Biol. 1996, 260, 446–466. [Google Scholar] [CrossRef] [PubMed]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy; Springer: Berlin, Germany, 2007. [Google Scholar]
- Meera, K.B.; Debra, A.K. Fluorescence spectroscopy of soluble E. coli SPase I Δ2-75 reveals conformational changes in response to ligand binding. Proteins 2014, 82, 596–606. [Google Scholar]
- Haiyuan, D.; Ishita, M.; Donald, O. Lipid and Signal Peptide-Induced Conformational Changes within the C-Domain of Escherichia coli SecA Protein. Biochemistry 2001, 40, 1835–1843. [Google Scholar]
- Imhof, N.; Kuhn, A.; Gerken, U. Substrate-Dependent Conformational Dynamics of the Escherichia coli Membrane Insertase YidC. Biochemistry 2011, 50, 3229–3239. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.F.; McCammon, A.J. Dynamics and design of enzymes and inhibitors. J. Am. Chem. Soc. 1986, 108, 3830–3832. [Google Scholar] [CrossRef]
- Liu, X.; Cao, Y.F.; Ran, R.X.; Dong, P.P.; Gonzalez, F.J.; Wu, X.; Huang, T.; Chen, J.X.; Fu, Z.W.; Li, R.S.; et al. New insights into the risk of phthalates: Inhibition of UDP-glucuronosyltransferases. Chemosphere 2016, 144, 1966–1972. [Google Scholar] [CrossRef] [PubMed]
- Cote, J.M.; Ramirez-Mondragon, C.A.; Siegel, Z.S.; Czyzyk, D.J.; Gao, J.; Sham, Y.Y.; Mukerji, I.; Taylor, E.A. The Stories Tryptophans Tell: Exploring Protein Dynamics of Heptosyltransferase I from Escherichia coli. Biochemistry 2017, 56, 886–895. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Peng, K.C.; Chen, L.; Puett, D.; Pierce, M. Circular Dichroic Spectroscopy of N-Acetylglucosaminyltransferase V and Its Substrate Interactions. J. Biol. Chem. 1997, 272, 4225–4229. [Google Scholar] [CrossRef] [PubMed]
- Cavatorta, P.; Sartor, G.; Neyroz, P.; Farruggia, C.; Franzoni, L.; Szabo, A.G.; Spisni, A. Fluorescence and CD studies on the conformation of the gastrin releasing peptide in solution and in the presence of model membranes. Biopolymers 1991, 31, 653–661. [Google Scholar] [CrossRef] [PubMed]












© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cote, J.M.; Taylor, E.A. The Glycosyltransferases of LPS Core: A Review of Four Heptosyltransferase Enzymes in Context. Int. J. Mol. Sci. 2017, 18, 2256. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18112256
Cote JM, Taylor EA. The Glycosyltransferases of LPS Core: A Review of Four Heptosyltransferase Enzymes in Context. International Journal of Molecular Sciences. 2017; 18(11):2256. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18112256
Chicago/Turabian StyleCote, Joy M., and Erika A. Taylor. 2017. "The Glycosyltransferases of LPS Core: A Review of Four Heptosyltransferase Enzymes in Context" International Journal of Molecular Sciences 18, no. 11: 2256. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18112256