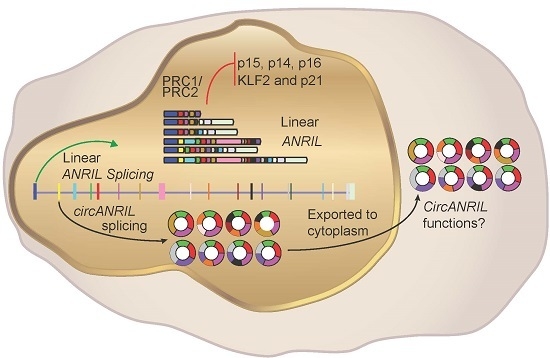
Multiple Isoforms of ANRIL in Melanoma Cells: Structural Complexity Suggests Variations in Processing
Abstract
:1. Introduction
2. Results
2.1. Differential Expression of ANRIL Exons in Melanoma Lines
2.2. Expression and Stability of circANRIL Relative to Linear ANRIL
2.3. Characterisation of circANRIL Isoforms
2.4. Non-Canonical Back-Splicing of ANRIL
2.5. Subcellular Localisation of Linear and Circular Isoforms of ANRIL Transcript
3. Discussion
3.1. Existence of Multiple Isoforms of ANRIL—Linear and Circular
3.2. Diversity of CircANRIL Species in Melanoma
3.3. Back-Spliced Junctions in CircANRIL Species: Not Mediated by Intronic Alu Repeats
3.4. Localisation of Circular and Linear ANRIL
4. Materials and Methods
4.1. Culture Conditions of Melanoma Lines
4.2. RNA Isolation and cDNA Synthesis
4.3. Reverse Transcription-PCR (RT-PCR)
4.4. Quantitative PCR (qPCR)
4.5. Gene Expression and TCGA Data Analysis
4.6. PCR and Sequencing of the Exon 14-5 Junction of circANRIL
4.7. Stability Assay for Circular and Linear Isoforms of ANRIL
4.8. Characterisation of circANRIL Isoform Using Outward Facing Primer
4.9. Intron and Exon Repeat Elements Analysis
4.10. Exon Skipping Alternative Splicing Analysis
4.11. Cell Fractionation
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pasmant, E.; Laurendeau, I.; Heron, D.; Vidaud, M.; Vidaud, D.; Bieche, I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: Identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007, 67, 3963–3969. [Google Scholar] [CrossRef] [PubMed]
- Zhang, E.B.; Kong, R.; Yin, D.D.; You, L.H.; Sun, M.; Han, L.; Xu, T.P.; Xia, R.; Yang, J.S.; De, W.; et al. Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR-99a/miR-449a. Oncotarget 2014, 5, 2276–2292. [Google Scholar] [CrossRef] [PubMed]
- Meseure, D.; Vacher, S.; Alsibai, K.D.; Nicolas, A.; Chemlali, W.; Caly, M.; Lidereau, R.; Pasmant, E.; Callens, C.; Bieche, I. Expression of ANRIL-polycomb complexes-CDKN2A/B/ARF genes in breast tumors: Identification of a two-gene (EZH2/CBX7) signature with independent prognostic value. Mol. Cancer Res. 2016, 14, 623–633. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.H.; Kim, D.; Jin, E.J. Down-regulation of phospholipase D stimulates death of lung cancer cells involving up-regulation of the long ncRNA ANRIL. Anticancer Res. 2015, 35, 2795–2803. [Google Scholar] [PubMed]
- Zhu, H.; Li, X.; Song, Y.; Zhang, P.; Xiao, Y.; Xing, Y. Long non-coding RNA ANRIL is up-regulated in bladder cancer and regulates bladder cancer cell proliferation and apoptosis through the intrinsic pathway. Biochem. Biophys. Res. Commun. 2015, 467, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Congrains, A.; Kamide, K.; Ohishi, M.; Rakugi, H. ANRIL: Molecular mechanisms and implications in human health. Int. J. Mol. Sci. 2013, 14, 1278–1292. [Google Scholar] [CrossRef] [PubMed]
- Yap, K.L.; Li, S.; Muñoz-Cabello, A.M.; Raguz, S.; Zeng, L.; Mujtaba, S. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 2010, 38, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene 2011, 30, 1956–1962. [Google Scholar] [CrossRef] [PubMed]
- Folkersen, L.; Kyriakou, T.; Goel, A.; Peden, J.; Malarstig, A.; Paulsson-Berne, G.; Hamsten, A.; Franco-Cereceda, A.; Gabrielsen, A.; Eriksson, P. Relationship between CAD risk genotype in the chromosome 9p21 locus and gene expression. Identification of eight new ANRIL splice variants. PLoS ONE 2009, 4, e7677. [Google Scholar] [CrossRef] [PubMed]
- Burd, C.E.; Jeck, W.R.; Liu, Y.; Sanoff, H.K.; Wang, Z.; Sharpless, N.E. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010, 6, e1001233. [Google Scholar] [CrossRef] [PubMed]
- Cunnington, M.S.; Koref, M.S.; Mayosi, B.M.; Burn, J.; Keavney, B. Chromosome 9p21 SNPs associated with multiple disease phenotypes correlate with ANRIL expression. PLoS Genet 2010, 6, e1000899. [Google Scholar] [CrossRef] [PubMed]
- Congrains, A.; Kamide, K.; Oguro, R.; Yasuda, O.; Miyata, K.; Yamamoto, E.; Takeya, Y.; Yamamoto, K. Genetic variants at the 9p21 locus contribute to atherosclerosis through modulation of ANRIL and CDKN2A/B. Atherosclerosis 2012, 220, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Davidovich, C.; Wang, X.; Cifuentes-Rojas, C.; Goodrich, K.J.; Gooding, A.R.; Lee, J.T.; Cech, T.R. Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol. Cell 2015, 57, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Holdt, L.M.; Hoffmann, S.; Sass, K.; Langenberger, D.; Scholz, M.; Krohn, K.; Finstermeier, K.; Stahringer, A.; Wilfert, W.; Beutner, F.; et al. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 2013, 9, e1003588. [Google Scholar] [CrossRef] [PubMed]
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016, 7, 12429. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sorrentino, J.A.; Wang, K.; Slevin, M.K.; Burd, C.E.; Liu, J.; Marzluff, W.F.; Sharpless, N.E. Circular RNAs are abundant, conserved, and associated with Alu repeats. RNA 2013, 19, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Rachakonda, P.S.; Heidenreich, B.; Nagore, E.; Sucker, A; Hemminki, K.; Schadendorf, D.; Kumar, R. Mapping of deletion breakpoints at the CDKN2A locus in melanoma: Detection of MTAP-ANRIL fusion transcripts. Oncotarget 2016, 7, 16490–16504. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, J.E. Long noncoding RNAs: Re-writing dogmas of RNA processing and stability. Biochim. Biophys. Acta 2015, 1859, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotech. 2014, 32, 453–461. [Google Scholar] [CrossRef] [PubMed]
- Cabili, M.N.; Dunagin, M.C.; McClanahan, P.D.; Biaesch, A.; Padovan-Merhar, O.; Regev, A.; Rinn, J.L.; Raj, A.; et al. Localization and abundance analysis of human lncRNAs at single-cell and single-molecule resolution. Genome Biol. 2015, 16, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Van Heesch, S.; van Iterson, M.; Jacobi, J.; Boymans, S.; Essers, P.; de Bruijn, E.; Hao, W.; MacInnes, A.W.; Cuppen, E.; Simonis, M. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol. 2014, 15, R6. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Dehn, J.; Droop, J.; Niegisch, G.; Niedworok, C.; Szarvas, T.; Schulz, W. Truncated Isoforms of lncRNA ANRIL are overexpressed in bladder cancer, but do not contribute to repression of INK4 tumor suppressors. Non-Coding RNA 2015, 1, 266. [Google Scholar] [CrossRef]
- Zhou, X.; Han, X.; Wittfeldt, A.; Sun, J.; Liu, C.; Wang, X.; Gan, L.M.; Cao, H.; Liang, Z. Long non-coding RNA ANRIL regulates inflammatory responses as a novel component of NF-κB pathway. RNA Biol. 2016, 13, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Wan, G.; Mathur, R.; Hu, X.; Liu, Y.; Zhang, X.; Peng, G. Long non-coding RNA ANRIL (CDKN2B-AS) is induced by the ATM-E2F1 signaling pathway. Cell Signal. 2013, 25, 1086–1095. [Google Scholar] [CrossRef] [PubMed]
- Nie, F.Q.; Sun, M.; Yang, J.S.; Xie, M.; Xu, T.P.; Xia, R.; Liu, Y.W.; Liu, X.H.; Zhang, E.B.; Lu, K.H.; et al. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol. Cancer Ther. 2015, 14, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Decatur, C.L.; Ong, E.; Garg, N.; Anbunathan, H.; Bowcock, A.M.; Field, M.G.; Harbour, J.W. Driver mutations in uveal melanoma: Associations with gene expression profile and patient outcomes. JAMA Ophthalmol. 2016, 134, 728–733. [Google Scholar] [CrossRef] [PubMed]
- Alsafadi, S.; Houy, A.; Battistella, A.; Popova, T.; Wassef, M.; Henry, E.; Tirode, F.; Constantinou, A.; Piperno-Neumann, S.; Roman-Roman, S.; et al. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat. Commun. 2016, 7, 10615. [Google Scholar] [CrossRef] [PubMed]
- Griewank, K.G.; Muller, H.; Jackett, L.A.; Emberger, M.; Moller, I.; van de Nes, J.A.P.; Zimmer, L.; Livingstone, E.; Wiesner, T.; Scholz, S.L.; et al. SF3B1 and BAP1 mutations in blue nevus-like melanoma. Mod. Pathol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Bullock, N.; Oltean, S. The many faces of SRPK1. J. Pathol. 2017, 241, 437–440. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Roychowdhury-Saha, M.; Black, C.; Watt, A.T.; Marcusson, E.G.; Freier, S.M.; Edgington, T.S. Control of RNA processing by a large non-coding RNA over-expressed in carcinomas. FEBS Lett. 2011, 585, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Wang, R.; Li, X.; Fan, M.; Lin, J.; Zhen, J.; Chen, L.; Lv, Z. LncRNA MALAT1 is dysregulated in diabetic nephropathy and involved in high glucose-induced podocyte injury via its interplay with β-catenin. J. Cell. Mol. Med. 2017. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Cheng, H.; Wang, G.; Yu, G.; Zhang, D.; Wang, Y.; Fan, W.; Yang, W. Deregulation of miR-183 promotes melanoma development via lncRNA MALAT1 regulation and ITGB1 signal activation. Oncotarget 2017, 8, 3509–3518. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Zhang, X.; Hao, Y.; Fang, Z.; He, Y. Potential roles of abnormally expressed long noncoding RNA UCA1 and Malat-1 in metastasis of melanoma. Melanoma Res. 2014, 24, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Marshall, E.S.; Matthews, J.H.; Shaw, J.H.; Nixon, J.; Tumewu, P.; Finlay, G.J.; Holdaway, K.M.; Baguley, B.C. Radiosensitivity of new and established human melanoma cell lines: Comparison of [3H]thymidine incorporation and soft agar clonogenic assays. Eur. J. Cancer 1994, 30, 1370–1376. [Google Scholar] [CrossRef]
- Glazar, P.; Papavasileiou, P.; Rajewsky, N. CircBase: A database for circular RNAs. RNA 2014, 20, 1666–1670. [Google Scholar] [CrossRef] [PubMed]
- R. Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2011. [Google Scholar]
- Alamancos, G.P.; Pagès, A.; Trincado, J.L.; Eyras, E. Leveraging transcript quantification for fast computation of alternative splicing profiles. RNA 2015, 21, 1521–1531. [Google Scholar] [CrossRef] [PubMed]





| Target Exons for Outward Primers | NZM7 CircANRIL Isoforms | NZM37 CircANRIL Isoforms |
|---|---|---|
| Exon 2 | 2-5-6-2 | |
| Exon 4 | 4-5-6-9-10-4 (10-4) 4-5-6-7-10-12-4 (12-4) 4-5-6-7-4 (7-4) 4-5-6-13-14-4 (14-4) 4-5-6-10-13-14-4 (14-4) 4-5-13-14-4 (14-4) 4-5-6-12-13-14-4 (14-4) 4-5-6-13-14-4 (14-4) 4-5-6-4 (6-4) 4-5-6-10-11-12-4 (12-4) | 4-5-6-7-4 (7-4) 4-5-6-7-13-14-4 (14-4) |
| Exon 6 | 6-4-5-6 (6-4) 6-14-5-6 (14-5) 6-7-9-10-5-6 (10-5) 6-9-10-5-6 (10-5) 6-10-2-5-6 (10-2) 6-4(N1)-4(N2)-5-6 (6-4N1) 6-4(N2)-4-5-6 (6-4N2) 6-7-5-6 (7-5) 6-10-5-6 (10-5) 6-10-4-5-6 (10-4) | 6-7-10-4-5-6 (10-4) 6-7-10-5-6 (10-5) 6-14-5-6 (14-5) 6-7-9-10-5-6 (10-5) 6-4-5-6 (6-4) |
| Exon 7 | 7-5-6-7 (7-5) | |
| Exon 8 | 8-5-6-8 (8-5) 8-5-6-7-8 (8-5) 8-9-10-5-6-7-8 (10-5) 8-13-14-5-6-8 (14-5) 8-10-13-14-5-6-8 (14-5) | |
| Exon 14 | 14-4-5-6-7-14 (14-4) 14-4-5-6-14 (14-4) 14-5-6-13N1-13-14 (14-5) 14-5-6-7-13-14 (14-5) 14-16-13N1-13-14 (16-13N1) | 14-5-6-14 (14-5) 14-5-6-13-14 (14-5) 14-4-5-6-7-14 (14-4) 14-5-6-7-13-14 (14-5) 14-4-5-6-7-9-14 (14-4) 14-4-5-6-7-13-14 (14-4) 14-5-6-7-10-13-14 (14-5) 14-5-6-7-9-10-13-14 (14-5) 14-4-5-6-7-10-13-14 (14-4) 14-5-6-7-10-12-13-14 (14-5) |
| Exon 16 | 16-15-16 (16-15) 16-5-6-7-13-14-15-16 (16-5) 16-6-7-13-14-15-16 (16-4) 16-19-5-6-10-13-14-15-16 (19-5) 16-4-5-6-7-13-14-15-16 (16-4) |
| Exon–Intron Junction | Potential CircANRIL |
|---|---|
| CDKN2B-AS1;SE:chr9:22029593-22032673:22032985-22046316:+ | 3 |
| CDKN2B-AS1;SE:chr9:22049227-22056251:22056386-22077678:+ | 7, 8, 9, 10, 11, 12 |
| CDKN2B-AS1;SE:chr9:22049227-22056251:22056386-22112319:+ CDKN2B-AS1;SE:chr9:22049227-22097257:22097363-22112319:+ | 7, 8, 9, 10, 11, 12, 13, 14 |
| CDKN2B-AS1;SE:chr9:22049227-22056251:22056386-22120199:+ | 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17 |
| CDKN2B-AS1;SE:chr9:22049227-22120199:22120409-22120503:+ | 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18 |
| CDKN2B-AS1;SE:chr9:22056386-22058358:22059053-22061952:+ | 8 |
| CDKN2B-AS1;SE:chr9:22056386-22061952:22062025-22063943:+ | 8, 9 |
| CDKN2B-AS1;SE:chr9:22056386-22063943:22064017-22077678:+ | 8, 9, 10, 11, 12 |
| CDKN2B-AS1;SE:chr9:22064017-22065661:22065756-22066234:+ | 11 |
| CDKN2B-AS1;SE:chr9:22064017-22066234:22066352-22077678:+ | 11, 12 |
| CDKN2B-AS1;SE:chr9:22112394-22113665:22113798-22118643:+ | 16 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarkar, D.; Oghabian, A.; Bodiyabadu, P.K.; Joseph, W.R.; Leung, E.Y.; Finlay, G.J.; Baguley, B.C.; Askarian-Amiri, M.E. Multiple Isoforms of ANRIL in Melanoma Cells: Structural Complexity Suggests Variations in Processing. Int. J. Mol. Sci. 2017, 18, 1378. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18071378
Sarkar D, Oghabian A, Bodiyabadu PK, Joseph WR, Leung EY, Finlay GJ, Baguley BC, Askarian-Amiri ME. Multiple Isoforms of ANRIL in Melanoma Cells: Structural Complexity Suggests Variations in Processing. International Journal of Molecular Sciences. 2017; 18(7):1378. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18071378
Chicago/Turabian StyleSarkar, Debina, Ali Oghabian, Pasani K. Bodiyabadu, Wayne R. Joseph, Euphemia Y. Leung, Graeme J. Finlay, Bruce C. Baguley, and Marjan E. Askarian-Amiri. 2017. "Multiple Isoforms of ANRIL in Melanoma Cells: Structural Complexity Suggests Variations in Processing" International Journal of Molecular Sciences 18, no. 7: 1378. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18071378