MicroRNA Signature of Human Microvascular Endothelium Infected with Rickettsia rickettsii
Abstract
:1. Introduction
2. Results
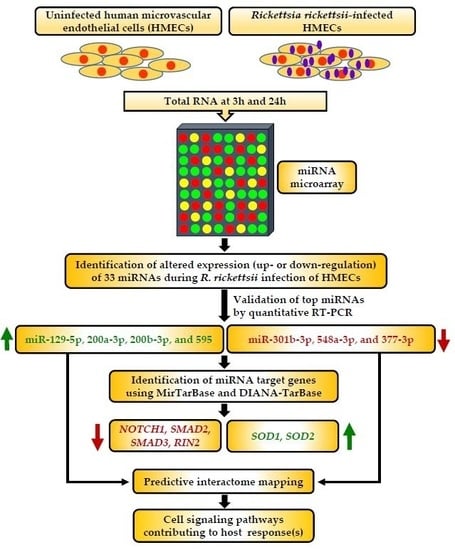
2.1. MicroRNA Expression Profiling of Microvascular Endothelial Cells Infected with R. rickettsii
2.2. Confirmatory Analysis of Differentially Regulated miRNAs Using In Vitro and In Vivo Models of Infection
2.3. Bioinformatics-Based Prediction of mRNA Targets for Differentially Regulated miRNAs
2.4. Correlation between Expression Profiles for miRNA-Target mRNA Pairs
2.5. miRNA200a-3p Controls NOTCH1 in R. rickettsii-Infected Endothelial Cells
2.6. Potential Regulatory Roles of miRNAs and Target mRNAs in Signaling Networks
3. Discussion
4. Materials and Methods
4.1. Endothelial Cell Culture, Infection, and Transfection
4.2. RNA Preparation
4.3. MicroRNA Expression Analysis by Affymetrix GeneChip® Microarray
4.4. Quantitative Real Time PCR for Analysis of miRNA and mRNA Expression
4.5. Identification of Target Genes
4.6. In Vivo Model of Infection
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Clifton, D.R.; Rydkina, E.; Huyck, H.; Pryhuber, G.; Freeman, R.S.; Silverman, D.J.; Sahni, S.K. Expression and secretion of chemotactic cytokines IL-8 and MCP-1 by human endothelial cells after Rickettsia rickettsii infection: Regulation by nuclear transcription factor NF-κB. Int. J. Med. Microbiol. 2005, 295, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Sporn, L.A.; Marder, V.J. Interleukin-1α production during Rickettsia rickettsii infection of cultured endothelial cells: potential role in autocrine cell stimulation. Infect. Immun. 1996, 64, 1609–1613. [Google Scholar] [PubMed]
- Valbuena, G.; Walker, D.H. Expression of CX3CL1 (fractalkine) in mice with endothelial-target rickettsial infection of the spotted-fever group. Virchows. Arch. 2005, 446, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Kaplanski, G.; Teysseire, N.; Farnarier, C.; Kaplanski, S.; Lissitzky, J.C.; Durand, J.M.; Soubeyrand, J.; Dinarello, C.A.; Bongrand, P. IL-6 and IL-8 production from cultured human endothelial cells stimulated by infection with Rickettsia conorii via a cell-associated IL-1α-dependent pathway. J. Clin. Investig. 1995, 96, 2839–2844. [Google Scholar] [CrossRef] [PubMed]
- Sahni, A.; Narra, H.P.; Walker, D.H.; Sahni, S.K. Endothelial activation and injury: The mechanisms of rickettsial vasculitis. In Vascular Responses to Pathogens; Gavins, F.N.E., Stokes, K.Y., Eds.; Elsevier Press: Atlanta, GA, USA, 2016; pp. 111–122. [Google Scholar]
- Schroeder, C.L.; Chowdhury, I.H.; Narra, H.P.; Patel, J.; Sahni, A.; Sahni, S.K. Human rickettsioses: Host response and molecular pathogenesis. In Rickettsiales: Biology, Molecular Biology, Epidemiology and Vaccine Development; Thomas, S., Ed.; Springer: New York, NY, USA, 2016; pp. 399–446. [Google Scholar]
- Sahni, S.K.; Narra, H.P.; Sahni, A.; Walker, D.H. Recent molecular insights into rickettsial pathogenesis and immunity. Future Microbiol. 2013, 8, 1265–1288. [Google Scholar] [CrossRef] [PubMed]
- Carrington, J.C.; Ambros, V. Role of microRNAs in plant and animal development. Science 2003, 301, 336–338. [Google Scholar] [CrossRef] [PubMed]
- Teleman, A.A.; Maitra, S.; Cohen, S.M. Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes Dev. 2006, 20, 417–422. [Google Scholar] [CrossRef] [PubMed]
- Borsani, O.; Zhu, J.; Verslues, P.E.; Sunkar, R.; Zhu, J.K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005, 123, 1279–1291. [Google Scholar] [CrossRef] [PubMed]
- Naeem, A.; Zhong, K.; Moisa, S.J.; Drackley, J.K.; Moyes, K.M.; Loor, J.J. Bioinformatics analysis of microRNA and putative target genes in bovine mammary tissue infected with Streptococcus uberis. J. Dairy Sci. 2012, 95, 6397–6408. [Google Scholar] [CrossRef] [PubMed]
- Correia, C.N.; Nalpas, N.C.; McLoughlin, K.E.; Browne, J.A.; Gordon, S.V.; MacHugh, D.E.; Shaughnessy, R.G. Circulating microRNAs as potential biomarkers of infectious disease. Front. Immunol. 2017, 8, 118. [Google Scholar] [CrossRef] [PubMed]
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Berezikov, E.; Guryev, V.; van de Belt, J.; Wienholds, E.; Plasterk, R.H.; Cuppen, E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell 2005, 120, 21–24. [Google Scholar] [CrossRef] [PubMed]
- Jovanovic, M.; Hengartner, M.O. miRNAs and apoptosis: RNAs to die for. Oncogene 2006, 25, 6176–6187. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Chen, D.; He, J.; Cai, J.; Shen, K.; Liu, X.; Yang, X.; Xu, L. Hcmv-miR-UL112 attenuates NK cell activity by inhibition type I interferon secretion. Immunol. Lett. 2015, 163, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Stern-Ginossar, N.; Elefant, N.; Zimmermann, A.; Wolf, D.G.; Saleh, N.; Biton, M.; Horwitz, E.; Prokocimer, Z.; Prichard, M.; Hahn, G.; et al. Host immune system gene targeting by a viral miRNA. Science 2007, 317, 376–381. [Google Scholar] [CrossRef] [PubMed]
- Reddick, L.E.; Alto, N.M. Bacteria fighting back: How pathogens target and subvert the host innate immune system. Mol. Cell 2014, 54, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Valbuena, G.; Walker, D.H. Effect of blocking the CXCL9/10-CXCR3 chemokine system in the outcome of endothelial-target rickettsial infections. Am. J. Trop Med. Hyg. 2004, 71, 393–399. [Google Scholar] [PubMed]
- Thomson, D.W.; Bracken, C.P.; Goodall, G.J. Experimental strategies for microRNA target identification. Nucleic Acids Res. 2011, 39, 6845–6853. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Stark, A.; Russell, R.B.; Cohen, S.M. Principles of microRNA-target recognition. PLoS Biol. 2005, 3, e85. [Google Scholar] [CrossRef] [PubMed]
- Chou, C.H.; Chang, N.W.; Shrestha, S.; Hsu, S.D.; Lin, Y.L.; Lee, W.H.; Yang, C.D.; Hong, H.C.; Wei, T.Y.; Tu, S.J.; et al. miRTarBase 2016: Updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res. 2016, 44, D239–D247. [Google Scholar] [CrossRef] [PubMed]
- Vlachos, I.S.; Paraskevopoulou, M.D.; Karagkouni, D.; Georgakilas, G.; Vergoulis, T.; Kanellos, I.; Anastasopoulos, I.L.; Maniou, S.; Karathanou, K.; Kalfakakou, D.; et al. DIANA-TarBase v7.0: Indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015, 43, D153–D159. [Google Scholar] [CrossRef] [PubMed]
- Sahni, S.K.; Kiriakidi, S.; Colonne, M.P.; Sahni, A.; Silverman, D.J. Selective activation of signal transducer and activator of transcription (STAT) proteins STAT1 and STAT3 in human endothelial cells infected with Rickettsia rickettsii. Clin. Microbiol. Infect. 2009, 15 (Suppl. S2), 303–304. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.J.; Cossart, P. Early signaling events involved in the entry of Rickettsia conorii into mammalian cells. J. Cell Sci. 2004, 117 Pt 21, 5097–5106. [Google Scholar] [CrossRef] [PubMed]
- Serio, A.W.; Jeng, R.L.; Haglund, C.M.; Reed, S.C.; Welch, M.D. Defining a core set of actin cytoskeletal proteins critical for actin-based motility of Rickettsia. Cell Host Microbe 2010, 7, 388–398. [Google Scholar] [CrossRef] [PubMed]
- Reed, S.C.; Lamason, R.L.; Risca, V.I.; Abernathy, E.; Welch, M.D. Rickettsia actin-based motility occurs in distinct phases mediated by different actin nucleators. Curr. Biol. 2014, 24, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Walker, D.H.; Popov, V.L.; Wen, J.; Feng, H.M. Rickettsia conorii infection of C3H/HeN mice. A model of endothelial-target rickettsiosis. Lab. Investig. 1994, 70, 358–368. [Google Scholar] [PubMed]
- Valbuena, G.; Bradford, W.; Walker, D.H. Expression analysis of the T-cell-targeting chemokines CXCL9 and CXCL10 in mice and humans with endothelial infections caused by rickettsiae of the spotted fever group. Am. J. Pathol. 2003, 163, 1357–1369. [Google Scholar] [CrossRef]
- Santulli, G. MicroRNAs and Endothelial (Dys) function. J. Cell Physiol. 2016, 231, 1638–1644. [Google Scholar] [CrossRef] [PubMed]
- Rusca, N.; Monticelli, S. MiR-146a in immunity and disease. Mol. Biol. Int. 2011, 2011, 437301. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, I.; David, M. MicroRNAs in the immune response. Cytokine 2008, 43, 391–394. [Google Scholar] [CrossRef] [PubMed]
- Terrier, O.; Textoris, J.; Carron, C.; Marcel, V.; Bourdon, J.C.; Rosa-Calatrava, M. Host microRNA molecular signatures associated with human H1N1 and H3N2 influenza A viruses reveal an unanticipated antiviral activity for miR-146a. J. Gen. Virol. 2013, 94 Pt 5, 985–995. [Google Scholar] [CrossRef] [PubMed]
- Cameron, J.E.; Yin, Q.; Fewell, C.; Lacey, M.; McBride, J.; Wang, X.; Lin, Z.; Schaefer, B.C.; Flemington, E.K. Epstein-Barr virus latent membrane protein 1 induces cellular microRNA miR-146a, a modulator of lymphocyte signaling pathways. J. Virol. 2008, 82, 1946–1958. [Google Scholar] [CrossRef] [PubMed]
- Pichler, K.; Schneider, G.; Grassmann, R. MicroRNA miR-146a and further oncogenesis-related cellular microRNAs are dysregulated in HTLV-1-transformed T lymphocytes. Retrovirology 2008, 5, 100. [Google Scholar] [CrossRef] [PubMed]
- Clifton, D.R.; Rydkina, E.; Freeman, R.S.; Sahni, S.K. NF-κB activation during Rickettsia rickettsii infection of endothelial cells involves the activation of catalytic IκB kinases IKKα and IKKβ and phosphorylation-proteolysis of the inhibitor protein IkappaBalpha. Infect. Immun. 2005, 73, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.S.; Sivachandran, N.; Lau, A.; Boudreau, E.; Zhao, J.L.; Baltimore, D.; Delgado-Olguin, P.; Cybulsky, M.I.; Fish, J.E. MicroRNA-146 represses endothelial activation by inhibiting pro-inflammatory pathways. EMBO Mol. Med. 2013, 5, 1017–1034. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Becker Buscaglia, L.E.; Barker, J.R.; Li, Y. MicroRNAs in NF-kappaB signaling. J. Mol. Cell Biol. 2011, 3, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.H.; Chang, C.H.; Tsai, R.K.; Hong, Y.R.; Chuang, T.H.; Fan, K.T.; Peng, C.W.; Wu, C.Y.; Hsu, W.L.; Wang, L.S.; et al. Cross-regulation of proinflammatory cytokines by interleukin-10 and miR-155 in Orientia tsutsugamushi-infected human macrophages prevents cytokine storm. J. Investig. Dermatol. 2016, 136, 1398–1407. [Google Scholar] [CrossRef] [PubMed]
- Eremeeva, M.E.; Silverman, D.J. Rickettsia rickettsii infection of the EA.hy 926 endothelial cell line: Morphological response to infection and evidence for oxidative injury. Microbiology 1998, 144 Pt 8, 2037–2048. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.E.; Santucci, L.A.; Tian, X.; Silverman, D.J. Superoxide dismutase-dependent, catalase-sensitive peroxides in human endothelial cells infected by Rickettsia rickettsii. Infect. Immun. 1998, 66, 1293–1298. [Google Scholar] [PubMed]
- Devamanoharan, P.S.; Santucci, L.A.; Hong, J.E.; Tian, X.; Silverman, D.J. Infection of human endothelial cells by Rickettsia rickettsii causes a significant reduction in the levels of key enzymes involved in protection against oxidative injury. Infect. Immun. 1994, 62, 2619–2621. [Google Scholar] [PubMed]
- Santucci, L.A.; Gutierrez, P.L.; Silverman, D.J. Rickettsia rickettsii induces superoxide radical and superoxide dismutase in human endothelial cells. Infect. Immun. 1992, 60, 5113–5118. [Google Scholar] [PubMed]
- Kume, T. Novel insights into the differential functions of Notch ligands in vascular formation. J. Angiogenes Res. 2009, 1, 8. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, K.; Satoh, M.; Ii, M.; Silver, M.; Limbourg, F.P.; Mukai, Y.; Rikitake, Y.; Radtke, F.; Gridley, T.; Losordo, D.W.; et al. Critical role of endothelial Notch1 signaling in postnatal angiogenesis. Circ. Res. 2007, 100, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Li, X.; Tulpule, A.; Zhou, Y.; Scehnet, J.S.; Zhang, S.; Lee, J.S.; Chaudhary, P.M.; Jung, J.; Gill, P.S. KSHV-induced notch components render endothelial and mural cell characteristics and cell survival. Blood 2010, 115, 887–895. [Google Scholar] [CrossRef] [PubMed]
- Narayana, Y.; Balaji, K.N. NOTCH1 up-regulation and signaling involved in Mycobacterium bovis BCG-induced SOCS3 expression in macrophages. J. Biol. Chem. 2008, 283, 12501–12511. [Google Scholar] [CrossRef] [PubMed]
- Lina, T.T.; Dunphy, P.S.; Luo, T.; McBride, J.W. Ehrlichia chaffeensis TRP120 activates canonical Notch signaling to downregulate TLR2/4 expression and promote intracellular survival. MBio 2016, 7, e00672-16. [Google Scholar] [CrossRef] [PubMed]
- Gridley, T. Notch signaling in the vasculature. Curr. Top. Dev. Biol. 2010, 92, 277–309. [Google Scholar] [PubMed]
- Rydkina, E.; Turpin, L.C.; Sahni, S.K. Rickettsia rickettsii infection of human macrovascular and microvascular endothelial cells reveals activation of both common and cell type-specific host response mechanisms. Infect. Immun. 2010, 78, 2599–2606. [Google Scholar] [CrossRef] [PubMed]
- Narra, H.P.; Schroeder, C.L.; Sahni, A.; Rojas, M.; Khanipov, K.; Fofanov, Y.; Sahni, S.K. Small regulatory RNAs of Rickettsia conorii. Sci. Rep. 2016, 6, 36728. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, A.; Mueller, O.; Stocker, S.; Salowsky, R.; Leiber, M.; Gassmann, M.; Lightfoot, S.; Menzel, W.; Granzow, M.; Ragg, T. The RIN: An RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 2006, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, C.L.; Narra, H.P.; Sahni, A.; Rojas, M.; Khanipov, K.; Patel, J.; Shah, R.; Fofanov, Y.; Sahni, S.K. Identification and characterization of novel small RNAs in Rickettsia prowazekii. Front. Microbiol. 2016, 7, 859. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, C.L.; Narra, H.P.; Rojas, M.; Sahni, A.; Patel, J.; Khanipov, K.; Wood, T.G.; Fofanov, Y.; Sahni, S.K. Bacterial small RNAs in the Genus Rickettsia. BMC Genom. 2015, 16, 1075. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protocol. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]






| miRNA Name | Consensus Sequence | Fold Change | |
|---|---|---|---|
| Up Regulated miRNAs | 3 h | 24 h | |
| miR-129-5p | CUUUUUGCGGUCUGGGCUUGC | 19.8 | 9.5 |
| miR-200a-3p | UAACACUGUCUGGUAACGAUGU | 15.9 | 8.8 |
| miR-297 | AUGUAUGUGUGCAUGUGCAUG | 15.6 | 18.6 |
| miR-200b-3p | UAAUACUGCCUGGUAAUGAUGA | 14.7 | 10.2 |
| miR-595 | GAAGUGUGCCGUGGUGUGUCU | 10.4 | 9.3 |
| miR-574-5p | UGAGUGUGUGUGUGUGAGUGUGU | 7.2 | 8.6 |
| miR-647 | GUGGCUGCACUCACUUCCUUC | 4.1 | 5.0 |
| miR-615-3p | UCCGAGCCUGGGUCUCCCUCUU | 3.9 | 4.3 |
| miR-1224-5p | GUGAGGACUCGGGAGGUGG | 3.5 | 4.2 |
| miR-1238-3p | CUUCCUCGUCUGUCUGCCCC | 2.2 | 2.2 |
| miR-196a-5p | UAGGUAGUUUCAUGUUGUUGGG | 3.1 | - |
| miR-616-3p | AGUCAUUGGAGGGUUUGAGCAG | 3.9 | - |
| miR-760 | CGGCUCUGGGUCUGUGGGGA | 3.7 | - |
| miR-1275 | GUGGGGGAGAGGCUGUC | 2.1 | - |
| miR-146a-5p | UGAGAACUGAAUUCCAUGGGUU | - | 13.7 |
| miR-631 | AGACCUGGCCCAGACCUCAGC | - | 12.6 |
| miR-661 | UGCCUGGGUCUCUGGCCUGCGCGU | - | 7.7 |
| miR-766-3p | ACUCCAGCCCCACAGCCUCAGC | - | 4.6 |
| miR-1273a | GGGCGACAAAGCAAGACUCUUUCUU | - | 2.8 |
| miR-1202 | GUGCCAGCUGCAGUGGGGGAG | - | 2.7 |
| miR-943 | CUGACUGUUGCCGUCCUCCAG | - | 2.7 |
| miR-1250-5p | ACGGUGCUGGAUGUGGCCUUU | - | 2.6 |
| miR-1229-3p | CUCUCACCACUGCCCUCCCACAG | - | 2.5 |
| Down Regulated miRNAs | |||
| miR-301b-3p | CAGUGCAAUGAUAUUGUCAAAGC | −3.3 | −2.8 |
| miR-548a-3p | CAAAACUGGCAAUUACUUUUGC | −3.1 | −3.1 |
| miR-377-3p | AUCACACAAAGGCAACUUUUGU | −2.1 | −3.2 |
| miR-602 | GACACGGGCGACAGCUGCGGCCC | −2.1 | - |
| miR-802 | CAGUAACAAAGAUUCAUCCUUGU | −2.2 | - |
| miR-376b-3p | AUCAUAGAGGAAAAUCCAUGUU | - | −2.9 |
| miR-216b-5p | AAAUCUCUGCAGGCAAAUGUGA | - | −2.4 |
| miR-216a-5p | UAAUCUCAGCUGGCAACUGUGA | - | −2.4 |
| miR-410-3p | AAUAUAACACAGAUGGCCUGU | - | −2.3 |
| miR-29b-3p | UAGCACCAUUUGAAAUCAGUGUU | - | −2.1 |
| miRNAs | Validated Target Genes (miRTarBase and DIANA-TarBase) |
|---|---|
| miR-129-5p | CAMTA1, SOX4, NOTCH1, UBE2F, CDK6, BMPR2, GALNT1, FMR1 |
| miR-200a-3p | NOTCH1, CDK6, ZEB1, ZEB2, SMAD2, SMAD3, RIN2, VCAM1, KEAP1 |
| miR-200b-3p | ZEB1, ZEB2 SIP1, SMAD2, SMAD3, VEGFA, GATA4, KDR, XIAP, RIN2 |
| miR-595 | CALM1, TNIP2, PARD6A, ULK1, ACP1, SASH3, RAB10, HINT1 |
| miR-548a-3p | DSC2, FGF-7, CALM1, ILDR1, PRLR, TRIM13, PARP15, SOX4 |
| miR-377-3p | PAK1, SOD1, SOD2, SMAD5, PKD1, IGF2R, FKBP5, CARD8 |
| miR-301b-3p | OXA1L, FGFR1, SMAD4, LDLR, TGFBR2, SOX4, EDN1, SOD2 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sahni, A.; Narra, H.P.; Patel, J.; Sahni, S.K. MicroRNA Signature of Human Microvascular Endothelium Infected with Rickettsia rickettsii. Int. J. Mol. Sci. 2017, 18, 1471. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18071471
Sahni A, Narra HP, Patel J, Sahni SK. MicroRNA Signature of Human Microvascular Endothelium Infected with Rickettsia rickettsii. International Journal of Molecular Sciences. 2017; 18(7):1471. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18071471
Chicago/Turabian StyleSahni, Abha, Hema P. Narra, Jignesh Patel, and Sanjeev K. Sahni. 2017. "MicroRNA Signature of Human Microvascular Endothelium Infected with Rickettsia rickettsii" International Journal of Molecular Sciences 18, no. 7: 1471. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18071471