miR-206-3p Inhibits 3T3-L1 Cell Adipogenesis via the c-Met/PI3K/Akt Pathway
Abstract
:1. Introduction
2. Results
2.1. Expression Level of miR-206 during the Differentiation of 3T3-L1 Cells
2.2. Effect of miR-206 on the Adipogenic Differentiation of 3T3-L1 Cells
2.3. c-Met Restores the Inhibition Effect of miR-206
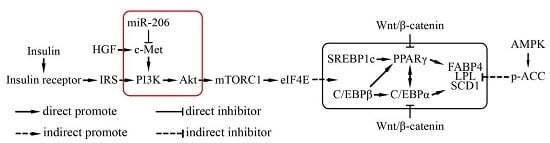
2.4. miR-206 Inactivates the c-Met/PI3k/Akt Pathway
3. Discussion
4. Materials and Methods
4.1. Silico Experiment
4.2. Cell Culture and Differentiation
4.3. Oil Red O Staining
4.4. Cell Transfection
4.5. Real-Time Quantitative Polymerase Chain Reaction
4.6. Western Blot
4.7. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| PPARγ | Peroxisome proliferator-activated receptor γ |
| C/EBPα | CCAAT/enhancer binding protein α |
| C/EBPβ | CCAAT/enhancer binding proteinβ |
| FABP4 | Fatty acid binding protein4 |
| PI3K | Phosphatidylinositol 3-kinase |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| HGF | Hepatocyte growth factor |
| HGFR | Hepatocyte growth factor receptor |
References
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Ambros, V. The functions of animal microRNAs. Nature 2004, 431, 350–355. [Google Scholar] [CrossRef] [PubMed]
- Tsujimoto, G. MicroRNAs, regulatory networks and diseases: An overview. FEBS J. 2011, 278, 1597. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Vienberg, S.; Geiger, J.; Madsen, S.; Dalgaard, L.T. MicroRNAs in metabolism. Acta Physiol. 2017, 219, 346–361. [Google Scholar] [CrossRef] [PubMed]
- Hilton, C.; Neville, M.J.; Karpe, F. MicroRNAs in adipose tissue: Their role in adipogenesis and obesity. Int. J. Obes. 2013, 37, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Yu, S.; Li, H.; Xiang, H.; Peng, J.; Jiang, S. MicroRNAs: Emerging roles in adipogenesis and obesity. Cell Signal. 2014, 26, 1888–1896. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, T.S.; Xu, Z.; Zhang, X.; Wang, L.; Gimble, J.M.; Lander, E.S.; Rosen, E.D. Comparative epigenomic analysis of murine and human adipogenesis. Cell 2010, 143, 156–169. [Google Scholar] [CrossRef] [PubMed]
- Townley-Tilson, W.H.; Callis, T.E.; Wang, D. MicroRNAs 1, 133, and 206: Critical factors of skeletal and cardiac muscle development, function, and disease. Int. J. Biochem. Cell Biol. 2010, 42, 1252–1255. [Google Scholar] [CrossRef] [PubMed]
- Walden, T.B.; Timmons, J.A.; Keller, P.; Nedergaard, J.; Cannon, B. Distinct expression of muscle-specific microRNAs (myomirs) in brown adipocytes. J. Cell. Physiol. 2009, 218, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.W.; Xi, X.Q.; Wu, J.; Wan, Y.Y.; Hui, H.X.; Cao, X.F. MicroRNA-206 attenuates tumor proliferation and migration involving the downregulation of NOTCH3 in colorectal cancer. Oncol. Rep. 2015, 33, 1402–1410. [Google Scholar] [CrossRef] [PubMed]
- Xue, D.; Yang, Y.; Liu, Y.; Wang, P.; Dai, Y.; Liu, Q.; Chen, L.; Shen, J.; Ju, H.; Li, Y.; et al. MicroRNA-206 attenuates the growth and angiogenesis in non-small cell lung cancer cells by blocking the 14–3–3zeta/STAT3/HIF-1α/VEGF signaling. Oncotarget 2016, 7, 79805–79813. [Google Scholar] [PubMed]
- Chen, Q.Y.; Jiao, D.M.; Wang, J.; Hu, H.; Tang, X.; Chen, J.; Mou, H.; Lu, W. miR-206 regulates cisplatin resistance and EMT in human lung adenocarcinoma cells partly by targeting MET. Oncotarget 2016, 7, 24510–24526. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.Y.; Jiao, D.M.; Wu, Y.Q.; Chen, J.; Wang, J.; Tang, X.L.; Mou, H.; Hu, H.Z.; Song, J.; Yan, J.; et al. miR-206 inhibits HGF-induced epithelial-mesenchymal transition and angiogenesis in non-small cell lung cancer via c-Met /PI3k/Akt/mTOR pathway. Oncotarget 2016, 7, 18247–18261. [Google Scholar] [CrossRef] [PubMed]
- Park, J.Y.; Kim, Y.; Im, J.A.; You, S.; Lee, H. Inhibition of adipogenesis by oligonol through Akt-mTOR inhibition in 3T3-L1 adipocytes. Evid. Based Complement. Altern. Med. 2014, 2014, 895272. [Google Scholar] [CrossRef] [PubMed]
- Mi, L.; Chen, Y.; Zheng, X.; Li, Y.; Zhang, Q.; Mo, D.; Yang, G. MicroRNA-139–5p Suppresses 3T3-L1 preadipocyte differentiation through notch and IRS1/PI3K/Akt insulin signaling pathways. J. Cell. Biochem. 2015, 116, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Yun, S.J.; Kim, E.K.; Tucker, D.F.; Kim, C.D.; Birnbaum, M.J.; Bae, S.S. Isoform-specific regulation of adipocyte differentiation by Akt/protein kinase Bα. Biochem. Biophys. Res. Commun. 2008, 371, 138–143. [Google Scholar] [CrossRef] [PubMed]
- White, H.M.; Acton, A.J.; Kamocka, M.M.; Considine, R.V. Hepatocyte growth factor regulates neovascularization in developing fat pads. Am. J. Physiol. Endocrinol. Metab. 2014, 306, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Trusolino, L.; Comoglio, P.M. Scatter-factor and semaphorin receptors: Cell signalling for invasive growth. Nat. Rev. Cancer 2002, 2, 289–300. [Google Scholar] [CrossRef] [PubMed]
- Mariani, M.; McHugh, M.; Petrillo, M.; Sieber, S.; He, S.; Andreoli, M.; Wu, Z.; Fiedler, P.; Scambia, G.; Shahabi, S.; et al. HGF/c-Met axis drives cancer aggressiveness in the neo-adjuvant setting of ovarian cancer. Oncotarget 2014, 5, 4855–4867. [Google Scholar] [CrossRef] [PubMed]
- Ntambi, J.M.; Young-Cheul, K. Adipocyte differentiation and gene expression. J. Nutr. 2000, 130, 3122S–3126S. [Google Scholar] [PubMed]
- Sarjeant, K.; Stephens, J.M. Adipogenesis. Cold Spring Harb. Perspect. Biol. 2012, 4, a008417. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Liu, Z.; Li, S.; Yang, C.; Xue, R.; Xi, Y.; Wang, L.; Wang, S.; He, Q.; Huang, J.; et al. Down-regulation of c-Met and Bcl2 by microRNA-206, activates apoptosis, and inhibits tumor cell proliferation, migration and colony formation. Oncotarget 2015, 6, 25533–25574. [Google Scholar] [CrossRef] [PubMed]
- Mataki, H.; Seki, N.; Chiyomaru, T.; Enokida, H.; Goto, Y.; Kumamoto, T.; Machida, K.; Mizuno, K.; Nakagawa, M.; Inoue, H. Tumor-suppressive microRNA-206 as a dual inhibitor of MET and EGFR oncogenic signaling in lung squamous cell carcinoma. Int. J. Oncol. 2015, 46, 1039–1050. [Google Scholar] [CrossRef] [PubMed]
- Dey, B.K.; Gagan, J.; Dutta, A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol. Cell. Biol. 2011, 31, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Hu, Y.; Song, G.; Hao, C.J.; Cui, Y.; Xia, H.F.; Ma, X. miR-206 regulates neural cells proliferation and apoptosis via Otx2. Cell. Physiol. Biochem. 2012, 29, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Alteri, A.; de Vito, F.; Messina, G.; Pompili, M.; Calconi, A.; Visca, P.; Mottolese, M.; Presutti, C.; Grossi, M. Cyclin D1 is a major target of miR-206 in cell differentiation and transformation. Cell Cycle 2013, 12, 3781–3790. [Google Scholar] [CrossRef] [PubMed]
- Yin, K.; Yin, W.; Wang, Y.; Zhou, L.; Liu, Y.; Yang, G.; Wang, J.; Lu, J. miR-206 suppresses epithelial mesenchymal transition by targeting TGF-β signaling in estrogen receptor positive breast cancer cells. Oncotarget 2016, 7, 24537–24548. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.K.; Qiao, H.Y.; Fu, M.H.; Li, G.; Li, W.B.; Chen, Z.; Wei, J.; Liang, B.S. miR-206 attenuates denervation-induced skeletal muscle atrophy in rats through regulation of satellite cell differentiation via TGF-β1, Smad3, and HDAC4 Signaling. Med. Sci. Monit. 2016, 22, 1161–1170. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Tong, Z.K.; Zhou, J.Y.; Yao, Y.K.; Zhang, S.M.; Zhou, J.Y. MicroRNA-206 inhibits the viability and migration of human lung adenocarcinoma cells partly by targeting MET. Oncol. Lett. 2016, 12, 1171–1177. [Google Scholar] [CrossRef] [PubMed]
- Zhong, D.; Huang, G.; Zhang, Y.; Zeng, Y.; Xu, Z.; Zhao, Y.; He, X.; He, F. MicroRNA-1 and microRNA-206 suppress LXRα-induced lipogenesis in hepatocytes. Cell Signal. 2013, 25, 1429–1437. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, S.; Marais, R.; Zhu, A.X. The role of signaling pathways in the development and treatment of hepatocellular carcinoma. Oncogene 2010, 29, 4989–5005. [Google Scholar] [CrossRef] [PubMed]
- Bertola, A.; Bonnafous, S.; Cormont, M.; Anty, R.; Tanti, J.F.; Tran, A.; Le Marchand-Brustel, Y.; Gual, P. Hepatocyte growth factor induces glucose uptake in 3T3-L1 adipocytes through A Gab1/phosphatidylinositol 3-kinase/Glut4 pathway. J. Biol. Chem. 2007, 282, 10325–10332. [Google Scholar] [CrossRef] [PubMed]
- Xiao, F.; Zuo, Z.; Cai, G.; Kang, S.; Gao, X.; Li, T. miRecords: An integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009, 37, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, R.J. Overview of vector design for mammalian gene expression. Mol. Biotechnol. 2000, 16, 151–160. [Google Scholar] [CrossRef]





© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, R.; Ma, F.; Li, W.; Ouyang, S.; Liu, Z.; Wu, J. miR-206-3p Inhibits 3T3-L1 Cell Adipogenesis via the c-Met/PI3K/Akt Pathway. Int. J. Mol. Sci. 2017, 18, 1510. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18071510
Tang R, Ma F, Li W, Ouyang S, Liu Z, Wu J. miR-206-3p Inhibits 3T3-L1 Cell Adipogenesis via the c-Met/PI3K/Akt Pathway. International Journal of Molecular Sciences. 2017; 18(7):1510. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18071510
Chicago/Turabian StyleTang, Renqiao, Feifei Ma, Wei Li, Shengrong Ouyang, Zhuo Liu, and Jianxin Wu. 2017. "miR-206-3p Inhibits 3T3-L1 Cell Adipogenesis via the c-Met/PI3K/Akt Pathway" International Journal of Molecular Sciences 18, no. 7: 1510. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18071510