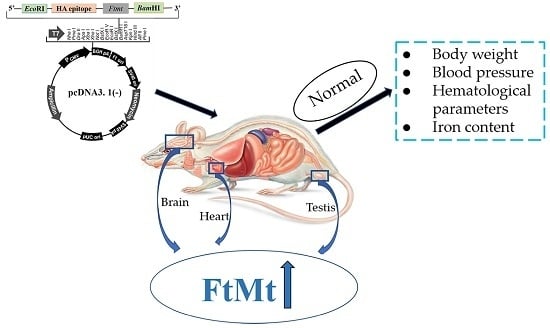
The Construction and Characterization of Mitochondrial Ferritin Overexpressing Mice
Abstract
:1. Introduction
2. Results
2.1. Generation of Transgenic Mice
2.2. Expression of FtMt is Increased in the Transgenic Mice
2.3. FtMt Overexpressing Mice Have Normal Body and Organ Weights
2.4. FtMt Overexpressing Mice Exhibit Normal Hematological Parameters and Blood Pressure
2.5. Transgenic Mice Have Normal Iron Distribution
2.6. The Effects of FtMt Overexpression on the Levels of Ferritin, TfR1, FPN1, and DMT1
2.7. The Locomotor Activity Was Not Changed in FtMt Overexpressing Mice
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. RNA Isolation and Real-Time PCR
4.3. Hematological Analyses
4.4. Blood Pressure and Heart Rate Measurement
4.5. Measurement of Serum and Tissue Iron
4.6. Measurement of Brain Iron
4.7. Western Blot Analysis
4.8. Locomotor Activity Test
4.9. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| FtMt | Mitochondrial ferritin |
| PD | Parkinson’s disease |
| AD | Alzheimer disease |
| ROS | Reactive oxygen species |
| IRE | Iron responsive element |
| IRP | Iron regulation protein |
| SEM | Standard error of the mean |
| RBC | Red blood cell count |
| HGB | Hemoglobin |
| HCT | Hematocrit |
| MCH | Mean corpuscular hemoglobin |
| MCHC | Mean corpuscular hemoglobin concentration |
| MCV | Mean corpuscular volume |
| RDW-CV | Coefficient variation of red blood cell volume distribution width |
| ICP-MS | Inductively coupled plasma mass spectrometry |
| FPN | Ferroportin |
| TfR1 | Transferrin receptor protein 1 |
| DMT1 | Divalent metal transporter 1 |
References
- Aisen, P.; Enns, C.; Wessling-Resnick, M. Chemistry and biology of eukaryotic iron metabolism. Int. J. Biochem. Cell Biol. 2001, 33, 940–959. [Google Scholar] [CrossRef]
- Hansen, J.B.; Moen, I.W.; Mandrup-Poulsen, T. Iron: The hard player in diabetes pathophysiology. Acta Physiol. 2014, 210, 717–732. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Chang, Y.Z. Mitochondrial ferritin in the regulation of brain iron homeostasis and neurodegenerative diseases. Front. Pharmacol. 2014, 5, 19. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Enns, C.A. Iron transport machinery of human cells: Players and their interactions. Curr. Top. Membr. 2012, 69, 67–93. [Google Scholar] [PubMed]
- Weiss, G.; Goodnough, L.T. Anemia of chronic disease. N. Engl. J. Med. 2005, 352, 1011–1023. [Google Scholar] [CrossRef] [PubMed]
- Fraenkel, P.G. Understanding anemia of chronic disease. Hematol. Am. Soc. Hematol. Educ. Program Book 2015, 2015, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Cullis, J. Anaemia of chronic disease. Clin. Med. 2013, 13, 193–196. [Google Scholar] [CrossRef] [PubMed]
- Lingor, P.; Carboni, E.; Koch, J.C. Alpha-synuclein and iron: Two keys unlocking Parkinson’s disease. J. Neural Transm. 2017. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wang, Z.Y. Metal ions influx is a double edged sword for the pathogenesis of Alzheimer’s disease. Ageing Res. Rev. 2017, 35, 265–290. [Google Scholar] [CrossRef] [PubMed]
- Gil-Lozano, C.; Davila, A.F.; Losa-Adams, E.; Fairen, A.G.; Gago-Duport, L. Quantifying fenton reaction pathways driven by self-generated H2O2 on pyrite surfaces. Sci. Rep. 2017, 7, 43703. [Google Scholar] [CrossRef] [PubMed]
- Levi, S.; Corsi, B.; Bosisio, M.; Invernizzi, R.; Volz, A.; Sanford, D.; Arosio, P.; Drysdale, J. A human mitochondrial ferritin encoded by an intronless gene. J. Biol. Chem. 2001, 276, 24437–24440. [Google Scholar] [CrossRef] [PubMed]
- Honarmand Ebrahimi, K.; Hagedoorn, P.L.; Hagen, W.R. Unity in the biochemistry of the iron-storage proteins ferritin and bacterioferritin. Chem. Rev. 2015, 115, 295–326. [Google Scholar] [CrossRef] [PubMed]
- Hagen, W.R.; Hagedoorn, P.L.; Honarmand Ebrahimi, K. The workings of ferritin: A crossroad of opinions. Metallomics 2017, 9, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Santambrogio, P.; Biasiotto, G.; Sanvito, F.; Olivieri, S.; Arosio, P.; Levi, S. Mitochondrial ferritin expression in adult mouse tissues. J. Histochem. Cytochem. 2007, 55, 1129–1137. [Google Scholar] [CrossRef] [PubMed]
- Levenson, C.W.; Tassabehji, N.M. Iron and ageing: An introduction to iron regulatory mechanisms. Ageing Res. Rev. 2004, 3, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Yang, H.; Guan, H.; Bellier, J.P.; Zhao, S.; Tooyama, I. Mapping of mitochondrial ferritin in the brainstem of Macaca fascicularis. Neuroscience 2016, 328, 92–106. [Google Scholar] [CrossRef] [PubMed]
- Corsi, B.; Cozzi, A.; Arosio, P.; Drysdale, J.; Santambrogio, P.; Campanella, A.; Biasiotto, G.; Albertini, A.; Levi, S. Human mitochondrial ferritin expressed in hela cells incorporates iron and affects cellular iron metabolism. J. Biol. Chem. 2002, 277, 22430–22437. [Google Scholar] [CrossRef] [PubMed]
- Nie, G.; Sheftel, A.D.; Kim, S.F.; Ponka, P. Overexpression of mitochondrial ferritin causes cytosolic iron depletion and changes cellular iron homeostasis. Blood 2005, 105, 2161–2167. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Yang, M.; Guan, H.; Liu, Z.; Zhao, S.; Takeuchi, S.; Yanagisawa, D.; Tooyama, I. Mitochondrial ferritin in neurodegenerative diseases. Neurosci. Res. 2013, 77, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yang, H.; Zhao, S.; Sato, H.; Konishi, Y.; Beach, T.G.; Abdelalim, E.M.; Bisem, N.J.; Tooyama, I. Expression and localization of mitochondrial ferritin mRNA in Alzheimer’s disease cerebral cortex. PLoS ONE 2011, 6, e22325. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.H.; Nie, G.; Duan, X.L.; Rouault, T.; Wu, W.S.; Ning, B.; Zhang, N.; Chang, Y.Z.; Zhao, B.L. Neuroprotective mechanism of mitochondrial ferritin on 6-hydroxydopamine-induced dopaminergic cell damage: Implication for neuroprotection in Parkinson’s disease. Antioxid. Redox Signal. 2010, 13, 783–796. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.S.; Zhao, Y.S.; Shi, Z.H.; Chang, S.Y.; Nie, G.J.; Duan, X.L.; Zhao, S.M.; Wu, Q.; Yang, Z.L.; Zhao, B.L.; et al. Mitochondrial ferritin attenuates β-amyloid-induced neurotoxicity: Reduction in oxidative damage through the Erk/P38 mitogen-activated protein kinase pathways. Antioxid. Redox Signal. 2013, 18, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wu, Q.; Wu, W.; Li, H.; Guo, Y.; Yu, P.; Gao, G.; Shi, Z.; Zhao, B.; Chang, Y.Z. Mitochondrial ferritin deletion exacerbates β-amyloid-induced neurotoxicity in mice. Oxid. Med. Cell. Longev. 2017, 2017, 1020357. [Google Scholar] [CrossRef] [PubMed]
- Doyle, A.; McGarry, M.P.; Lee, N.A.; Lee, J.J. The construction of transgenic and gene knockout/knockin mouse models of human disease. Transgenic Res. 2012, 21, 327–349. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Guan, H.; Yang, M.; Liu, Z.; Takeuchi, S.; Yanagisawa, D.; Vincent, S.R.; Zhao, S.; Tooyama, I. Upregulation of mitochondrial ferritin by proinflammatory cytokines: Implications for a role in Alzheimer’s disease. J. Alzheimer’s Dis. 2015, 45, 797–811. [Google Scholar]
- Camaschella, C. Understanding iron homeostasis through genetic analysis of hemochromatosis and related disorders. Blood 2005, 106, 3710–3717. [Google Scholar] [CrossRef] [PubMed]
- Hentze, M.W.; Muckenthaler, M.U.; Galy, B.; Camaschella, C. Two to tango: Regulation of mammalian iron metabolism. Cell 2010, 142, 24–38. [Google Scholar] [CrossRef] [PubMed]
- Hentze, M.W.; Muckenthaler, M.U.; Andrews, N.C. Balancing acts: Molecular control of mammalian iron metabolism. Cell 2004, 117, 285–297. [Google Scholar] [CrossRef]
- Brissot, P. Optimizing the diagnosis and the treatment of iron overload diseases. Expert Rev. Gastroenterol. Hepatol. 2016, 10, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Esposito, B.P.; Breuer, W.; Sirankapracha, P.; Pootrakul, P.; Hershko, C.; Cabantchik, Z.I. Labile plasma iron in iron overload: Redox activity and susceptibility to chelation. Blood 2003, 102, 2670–2677. [Google Scholar] [CrossRef] [PubMed]
- Cabantchik, Z.I.; Breuer, W.; Zanninelli, G.; Cianciulli, P. LPI-labile plasma iron in iron overload. Best Pract. Res. Clin. Haematol. 2005, 18, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, C.; Scapagini, G.; Curro, D.; Giuffrida Stella, A.M.; de Marco, C.; Butterfield, D.A.; Calabrese, V. Mitochondrial dysfunction, free radical generation and cellular stress response in neurodegenerative disorders. Front. Biosci. 2007, 12, 1107–1123. [Google Scholar] [CrossRef] [PubMed]
- Napier, I.; Ponka, P.; Richardson, D.R. Iron trafficking in the mitochondrion: Novel pathways revealed by disease. Blood 2005, 105, 1867–1874. [Google Scholar] [CrossRef] [PubMed]
- Della Porta, M.G.; Malcovati, L.; Invernizzi, R.; Travaglino, E.; Pascutto, C.; Maffioli, M.; Galli, A.; Boggi, S.; Pietra, D.; Vanelli, L.; et al. Flow cytometry evaluation of erythroid dysplasia in patients with myelodysplastic syndrome. Leukemia 2006, 20, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.Y.; Uprichard, J. Ferritin and iron studies in anaemia and chronic disease. Ann. Clin. Biochem. 2017, 54, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Waldvogel-Abramowski, S.; Waeber, G.; Gassner, C.; Buser, A.; Frey, B.M.; Favrat, B.; Tissot, J.D. Physiology of iron metabolism. Transfus. Med. Hemother. 2014, 41, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Arosio, P.; Carmona, F.; Gozzelino, R.; Maccarinelli, F.; Poli, M. The importance of eukaryotic ferritins in iron handling and cytoprotection. Biochem. J. 2015, 472, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Santambrogio, P.; Levi, S.; Cozzi, A.; Rovida, E.; Albertini, A.; Arosio, P. Production and characterization of recombinant heteropolymers of human ferritin H and L chains. J. Biol. Chem. 1993, 268, 12744–12748. [Google Scholar] [PubMed]
- Honarmand Ebrahimi, K.; Bill, E.; Hagedoorn, P.L.; Hagen, W.R. The catalytic center of ferritin regulates iron storage via Fe(II)-Fe(III) displacement. Nat. Chem. Biol. 2012, 8, 941–948. [Google Scholar] [CrossRef] [PubMed]
- Arosio, P.; Levi, S. Cytosolic and mitochondrial ferritins in the regulation of cellular iron homeostasis and oxidative damage. Biochim. Biophys. Acta 2010, 8, 783–792. [Google Scholar] [CrossRef] [PubMed]
- de la Pena, T.C.; Carcamo, C.B.; Diaz, M.I.; Brokordt, K.B.; Winkler, F.M. Molecular characterization of two ferritins of the scallop argopecten purpuratus and gene expressions in association with early development, immune response and growth rate. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2016, 198, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zak, O.; Aisen, P.; Harrison, S.C.; Walz, T. Structure of the human transferrin receptor-transferrin complex. Cell 2004, 116, 565–576. [Google Scholar] [CrossRef]
- Bartnikas, T.B.; Campagna, D.R.; Antiochos, B.; Mulhern, H.; Pondarre, C.; Fleming, M.D. Characterization of mitochondrial ferritin-deficient mice. Am. J. Hematol. 2010, 85, 958–960. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Chang, S.; Wu, Q.; Xu, Z.; Wang, P.; Li, Y.; Yu, P.; Gao, G.; Shi, Z.; Duan, X.; et al. Mitochondrial ferritin protects the murine myocardium from acute exhaustive exercise injury. Cell Death Dis. 2016, 7, e2475. [Google Scholar] [CrossRef] [PubMed]
- You, L.H.; Li, F.; Wang, L.; Zhao, S.E.; Wang, S.M.; Zhang, L.L.; Zhang, L.H.; Duan, X.L.; Yu, P.; Chang, Y.Z. Brain iron accumulation exacerbates the pathogenesis of MPTP-induced Parkinson’s disease. Neuroscience 2015, 284, 234–246. [Google Scholar] [CrossRef] [PubMed]
- You, L.H.; Yan, C.Z.; Zheng, B.J.; Ci, Y.Z.; Chang, S.Y.; Yu, P.; Gao, G.F.; Li, H.Y.; Dong, T.Y.; Chang, Y.Z. Astrocyte hepcidin is a key factor in lps-induced neuronal apoptosis. Cell Death Dis. 2017, 8, e2676. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yu, P.; Chang, S.Y.; Wu, Q.; Xie, C.; Wu, W.; Zhao, B.; Gao, G.; Chang, Y.Z. Hypobaric hypoxia regulates brain iron homeostasis in rats. J. Cell. Biochem. 2017, 118, 1596–1605. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.Z.; Qian, Z.M.; Wang, K.; Zhu, L.; Yang, X.D.; Du, J.R.; Jiang, L.; Ho, K.P.; Wang, Q.; Ke, Y. Effects of development and iron status on ceruloplasmin expression in rat brain. J. Cell. Physiol. 2005, 204, 623–631. [Google Scholar] [CrossRef] [PubMed]
- You, L.H.; Li, Z.; Duan, X.L.; Zhao, B.L.; Chang, Y.Z.; Shi, Z.H. Mitochondrial ferritin suppresses MPTP-induced cell damage by regulating iron metabolism and attenuating oxidative stress. Brain Res. 2016, 1, 33–42. [Google Scholar] [CrossRef] [PubMed]







© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Wang, P.; Wu, Q.; Xie, L.; Cui, Y.; Li, H.; Yu, P.; Chang, Y.-Z. The Construction and Characterization of Mitochondrial Ferritin Overexpressing Mice. Int. J. Mol. Sci. 2017, 18, 1518. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18071518
Li X, Wang P, Wu Q, Xie L, Cui Y, Li H, Yu P, Chang Y-Z. The Construction and Characterization of Mitochondrial Ferritin Overexpressing Mice. International Journal of Molecular Sciences. 2017; 18(7):1518. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18071518
Chicago/Turabian StyleLi, Xin, Peina Wang, Qiong Wu, Lide Xie, Yanmei Cui, Haiyan Li, Peng Yu, and Yan-Zhong Chang. 2017. "The Construction and Characterization of Mitochondrial Ferritin Overexpressing Mice" International Journal of Molecular Sciences 18, no. 7: 1518. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18071518