Investigating the Influence of Magnesium Ions on p53–DNA Binding Using Atomic Force Microscopy
Abstract
:1. Introduction
2. Results and Discussion
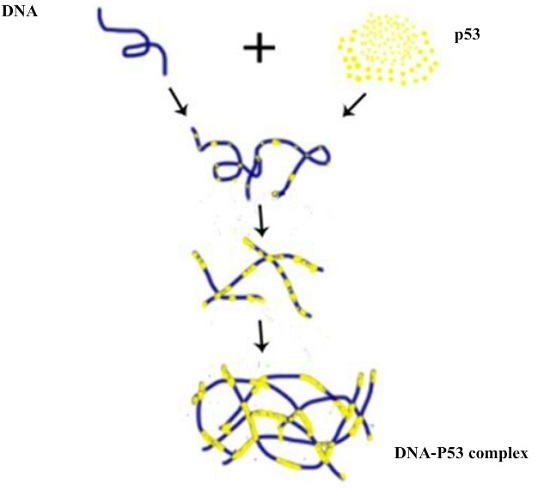
2.1. AFM Imaging of DNA and p53 Protein Samples
2.2. Dependence of p53–DNA Bindings on the Protein Concentration
2.3. p53–DNA Binding in the Influence of Magnesium Ions
3. Materials and Methods
3.1. DNA and Human p53 Protein
3.2. AFM Sample Preparation
3.2.1. DNA and p53 Samples
3.2.2. p53–DNA Complex Samples
3.3. AFM Imaging of p53–DNA Complexes
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Nuttall, P.; Lee, K.; Ciccarella, P.; Carminati, M.; Ferrari, G.; Kim, K.B.; Albrecht, T. Single-molecule studies of unlabeled full-length p53 protein binding to DNA. J. Phys. Chem. B 2016, 120, 2106–2114. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Cherny, D.I.; Heim, G.; Jovin, T.M.; Schäffer, T.E. Dynamic interactions of p53 with DNA in solution by time-lapse atomic force microscopy. J. Mol. Biol. 2001, 314, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.J.; Lain, S.; Verma, C.S.; Fersht, A.R.; Lane, D.P. Awakening guardian angels: Drugging the p53 pathway. Nat. Rev. Cancer 2009, 9, 862–873. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Lane, D.P.; Levine, A.J. Surfing the tp53 network. Nature 2000, 408, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Mirzayans, R.; Andrais, B.; Kumar, P.; Murray, D. The growing complexity of cancer cell response to DNA-damaging agents: Caspase 3 mediates cell death or survival? Int. J. Mol. Sci. 2016, 17, 708. [Google Scholar] [CrossRef] [PubMed]
- Mirzayans, R.; Andrais, B.; Kumar, P.; Murray, D. Significance of wild-type p53 signaling in suppressing apoptosis in response to chemical genotoxic agents: Impact on chemotherapy outcome. Int. J. Mol. Sci. 2017, 18, 928. [Google Scholar] [CrossRef] [PubMed]
- Chao, C.; Saito, S.I.; Anderson, C.W.; Appella, E.; Xu, Y. Phosphorylation of murine p53 at ser-18 regulates the p53 responses to DNA damage. Proc. Natl. Acad. Sci. USA 2000, 971, 11936–11941. [Google Scholar] [CrossRef] [PubMed]
- Cain, C.; Miller, S.; Ahn, J.; Prives, C. The n terminus of p53 regulates its dissociation from DNA. J. Biol. Chem. 2000, 275, 39944–39953. [Google Scholar] [CrossRef] [PubMed]
- Bargonetti, J.; Manfredi, J.J.; Chen, X.; Marshak, D.R.; Prives, C. A proteolytic fragment from the central region of p53 has marked sequence-specific DNA-binding activity when generated from wild-type but not from oncogenic mutant p53 protein. Genes Dev. 1993, 7, 2565–2574. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.J. P53, the cellular gatekeeper for growth and division. Cell 1997, 88, 323–331. [Google Scholar] [CrossRef]
- Stenger, J.E.; Tegtmeyer, P.; Mayr, G.A.; Reed, M.; Wang, Y.; Wang, P.; Hough, P.V.; Mastrangelo, I.A. P53 oligomerization and DNA looping are linked with transcriptional activation. EMBO J. 1994, 13, 6011–6020. [Google Scholar] [PubMed]
- Brázdová, M.; Paleček, J.; Cherny, D.I.; Billová, S.; Fojta, M.; Pečinka, P.; Vojtěšek, B.; Jovin, T.M.; Paleček, E. Role of tumor suppressor p53 domains in selective binding to supercoiled DNA. Nucleic Acids Res. 2002, 30, 4966–4974. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, C.; Murata, A.; Itoh, Y.; Subekti, D.R.G.; Takahashi, S.; Kamagata, K. DNA garden: A simple method for producing arrays of stretchable DNA for single-molecule fluorescence imaging of DNA binding proteins. Bull. Chem. Soc. Jpn. 2016, 90, 34–43. [Google Scholar] [CrossRef]
- Kamagata, K.; Murata, A.; Itoh, Y.; Takahashi, S. Characterization of facilitated diffusion of tumor suppressor p53 along DNA using single-molecule fluorescence imaging. J. Photoch. Photobio. C 2017, 30, 36–50. [Google Scholar] [CrossRef]
- Berg, O.G.; Blomberg, C. Association kinetics with coupled diffusion : III. Ionic-strength dependence of the lac repressor-operator association. Biophys. Chem. 1978, 8, 271–280. [Google Scholar] [CrossRef]
- Winter, R.B.; Von Hippel, P.H. Diffusion-driven mechanisms of protein translocation on nucleic acids. 2. The escherichia coli lac repressor-operator interaction: Equilibrium measurements. Biochemistry 1981, 20, 6948–6960. [Google Scholar] [PubMed]
- Terakawa, T.; Kenzaki, H.; Takada, S. p53 searches on DNA by rotation-uncoupled sliding at C-terminal tails and restricted hopping of core domains. J. Am. Chem. Soc. 2012, 134, 14555–14562. [Google Scholar] [CrossRef] [PubMed]
- Leith, J.; Tafvizi, A.; Huang, F.; Uspal, W.; Doyle, P.; Fersht, A.; Mirny, L.; Van Oijen, A. Sequence-dependent sliding kinetics of p53. Proc. Natl. Acad. Sci. USA 2012, 109, 16552–16557. [Google Scholar] [CrossRef] [PubMed]
- Tafvizi, A.; Mirny, L.A.; van Oijen, A.M. Dancing on DNA: Kinetic aspects of search processes on DNA. ChemPhysChem 2011, 12, 1481–1489. [Google Scholar] [CrossRef] [PubMed]
- Widom, J. Target site localization by site-specific, DNA-binding proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 16909–16910. [Google Scholar] [CrossRef] [PubMed]
- Melero, R.; Valle, M. Electron microscopy studies on the quaternary structure of p53 reveal different binding modes for p53 tetramers in complex with DNA. Proc. Natl. Acad. Sci. USA 2011, 108, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Halford, S.E.; Marko, J.F. How do site-specific DNA-binding proteins find their targets? Nucleic Acids Res. 2004, 32, 3040–3052. [Google Scholar] [CrossRef] [PubMed]
- Blainey, P.C.; van Oijen, A.M.; Banerjee, A.; Verdine, G.L.; Xie, X.S. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl. Acad. Sci. USA 2006, 103, 5752–5757. [Google Scholar] [CrossRef] [PubMed]
- Elf, J.; Li, G.W.; Xie, X.S. Probing transcription factor dynamics at the single-molecule level in a living cell. Science 2007, 316, 1191–1194. [Google Scholar] [CrossRef] [PubMed]
- Hu, T.; Grosberg, A.Y.; Shklovskii, B.I. How proteins search for their specific sites on DNA: The role of DNA conformation. Biophys. J. 2006, 90, 2731–2744. [Google Scholar] [CrossRef] [PubMed]
- Gowers, D.M.; Wilson, G.G.; Halford, S.E. Measurement of the contributions of 1d and 3d pathways to the translocation of a protein along DNA. Proc. Natl. Acad. Sci. USA 2005, 102, 15883. [Google Scholar] [CrossRef] [PubMed]
- Hainaut, P.; Milner, J. A structural role for metal ions in the “wild-type” conformation of the tumor suppressor protein p53. Cancer Res. 1993, 53, 1739–1742. [Google Scholar] [PubMed]
- Butler, J.S.; Loh, S.N. Structure, function, and aggregation of the zinc-free form of the p53 DNA binding domain. Biochemistry 2003, 42, 2396–2403. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Wang, S.X. Effect of metal ion on the structural stability of tumour suppressor protein p53 DNA-binding domain. J. Biochem. 2009, 146, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Wang, S.; Feng, X. Influence of magnesium ion on the binding of p53 DNA-binding domain to DNA-response elements. J. Biochem. 2009, 146, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Muir, J.K.; Raghupathi, R.; Emery, D.L.; Bareyre, F.M.; Mcintosh, T.K. Postinjury magnesium treatment attenuates traumatic brain injury-induced cortical induction of p53 mrna in rats. Exp. Neurol. 1999, 159, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Shao, X.; Cai, W.; Xue, Y.; Wang, S.; Feng, X. Predicting the coordination geometry for Mg2+ in the p53 DNA-binding domain: Insights from computational studies. Phys. Chem. Chem. Phys. 2011, 13, 1140–1151. [Google Scholar] [CrossRef] [PubMed]
- Drake, B.; Prater, C.B.; Weisenhorn, A.L.; Gould, S.A.C.; Albrecht, T.R.; Quate, C.F.; Cannell, D.S.; Hansma, H.G.; Hansma, P.K. Imaging crystals, polymers, and processes in water with the atomic force microscope. Science 1989, 243, 1586–1589. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, C.; Rivetti, C.; Keller, D.J. Scanning force microscopy under aqueous solutions. Curr. Opin. Struct. Biol. 1997, 7, 709–716. [Google Scholar] [CrossRef]
- Alessandrini, A.; Facci, P. Afm: A versatile tool in biophysics. Meas. Sci. Technol. 2005, 16, 65–92. [Google Scholar] [CrossRef]
- Wang, Y.; Ran, S.; Yang, G. Single molecular investigation of DNA looping and aggregation by restriction endonuclease bspmi. Sci. Rep. 2014, 4, 5897. [Google Scholar] [CrossRef] [PubMed]
- Cassina, V.; Seruggia, D.; Beretta, G.L.; Salerno, D.; Brogioli, D.; Manzini, S.; Zunino, F.; Mantegazza, F. Atomic force microscopy study of DNA conformation in the presence of drugs. Biophys. Struct. Mech. 2011, 40, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Leng, F.; Chen, B.; Dunlap, D.D. Dividing a supercoiled DNA molecule into two independent topological domains. Proc. Natl. Acad. Sci. USA 2011, 108, 19973–19978. [Google Scholar] [CrossRef] [PubMed]
- Murugesapillai, D. DNA bridging and looping by hmo1 provides a mechanism for stabilizing nucleosome-free chromatin. Nucleic Acids Res. 2014, 42, 8996–9004. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ran, S.; Man, B.; Yang, G. DNA condensations on mica surfaces induced collaboratively by alcohol and hexammine cobalt. Colloids Surf. B Biointerfaces 2011, 83, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ran, S.; Man, B.; Yang, G. Ethanol induces condensation of single DNA molecules. Soft Matter 2011, 7, 4425–4434. [Google Scholar] [CrossRef]
- Tafvizi, A.; Huang, F.; Fersht, A.R.; Mirny, L.A.; van Oijen, A.M. A single-molecule characterization of p53 search on DNA. Proc. Natl. Acad. Sci. USA 2011, 108, 563–568. [Google Scholar] [CrossRef] [PubMed]
- Murata, A.; Ito, Y.; Kashima, R.; Kanbayashi, S.; Nanatani, K.; Igarashi, C.; Okumura, M.; Inaba, K.; Tokino, T.; Takahashi, S. One-dimensional sliding of p53 along DNA is accelerated in the presence of Ca2+ or Mg2+ at millimolar concentrations. J. Mol. Biol. 2015, 427, 2663–2678. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Li, Z.; Zhou, H.; Wei, G.; Song, Y.; Wang, L. Imaging DNA molecules on mica surface by atomic force microscopy in air and in liquid. Microsc. Res. Tech. 2005, 66, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Tidow, H.; Melero, R.; Mylonas, E.; Freund, S.M.V.; Grossmann, J.G.; Carazo, J.M.; Svergun, D.I.; Valle, M.; Fersht, A.R. Quaternary structures of tumor suppressor p53 and a specific p53-DNA complex. Proc. Natl. Acad. Sci. USA 2007, 104, 12324–12329. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Guthold, M.; Snyder, M.J.; Lu, H. Afm of self-assembled lambda DNA-histone networks. Colloids Surf. B Biointerfaces 2015, 134, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Moomaw, A. S.; Maguire, M.E. The unique nature of Mg2+ channels. Physiology 2008, 23, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Qi, R.; Wei, G.; Nussinov, R.; Ma, B. Self-aggregation and coaggregation of the p53 core fragment with its aggregation gatekeeper variant. Phys. Chem. Chem. Phys. 2016, 18, 8098–8107. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.L.; Cv, D.M.G.; Costa, D.C.; Rangel, L.P. Prion-like aggregation of mutant p53 in cancer. Trends Biochem. Sci. 2014, 39, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Yang-Hartwich, Y.; Soteras, M.G.; Lin, Z.P.; Holmberg, J.; Sumi, N.; Craveiro, V.; Liang, M.; Romanoff, E.; Bingham, J.; Garofalo, F. p53 protein aggregation promotes platinum resistance in ovarian cancer. Oncogene 2015, 34, 3605–3616. [Google Scholar] [CrossRef] [PubMed]
- Nečas, D.; Klapetek, P. Gwyddion: An open-source software for spm data analysis. Cent. Eur. J. Phys. 2012, 10, 181–188. [Google Scholar] [CrossRef]






| The Concentration Ratio of p53 and DNA | p53 Molar Concentration (nmol/L) | The Quantity of p53 Adhering to DNA (/μm DNA) | Probability of DNA Binding Site Occupied by p53 |
|---|---|---|---|
| 0 | 0 | 0 | 0 |
| 1:1 | 18.867 | 0.8 ± 0.2 | 0.018 ± 0.005 |
| 1.5:1 | 28.3005 | 1.6 ± 0.5 | 0.036 ± 0.011 |
| 2:1 | 37.734 | 2.9 ± 0.6 | 0.064 ± 0.013 |
| 2.5:1 | 47.1675 | 4.9 ± 0.9 | 0.11 ± 0.021 |
| 3:1 | 56.601 | 10.2 ± 1.6 | 0.23 ± 0.037 |
| The Concentration Ratio of p53 and DNA | p53 Molar Concentration (nmol/L) | The Quantity of p53 Adhering to DNA(/μm DNA) | Probability of DNA Binding Site Occupied by p53 |
|---|---|---|---|
| 0 | 0 | 0 | 0 |
| 1:1 | 18.867 | 4.2 ± 1.8 | 0.093 ± 0.040 |
| 2:1 | 37.734 | 6.2 ± 4.9 | 0.137 ± 0.110 |
| 3:1 | 56.601 | 6.3 ± 3.8 | 0.139 ± 0.084 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.; Gao, T.; Wang, Y.; Yang, G. Investigating the Influence of Magnesium Ions on p53–DNA Binding Using Atomic Force Microscopy. Int. J. Mol. Sci. 2017, 18, 1585. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18071585
Chen Y, Gao T, Wang Y, Yang G. Investigating the Influence of Magnesium Ions on p53–DNA Binding Using Atomic Force Microscopy. International Journal of Molecular Sciences. 2017; 18(7):1585. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18071585
Chicago/Turabian StyleChen, Yang, Tianyong Gao, Yanwei Wang, and Guangcan Yang. 2017. "Investigating the Influence of Magnesium Ions on p53–DNA Binding Using Atomic Force Microscopy" International Journal of Molecular Sciences 18, no. 7: 1585. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18071585