Tumor-Stroma Crosstalk in Bone Tissue: The Osteoclastogenic Potential of a Breast Cancer Cell Line in a Co-Culture System and the Role of EGFR Inhibition
Abstract
:1. Introduction
2. Results
2.1. Mesenchymal stem cells (MSC) Induce the Expression of RANK and EGFR in Cancer Cells
2.2. Cancer Cells and MSC Contribute to Osteoclastogenesis
Cancer cells and COCO promote Osteoclastogenesis
2.3. Gef Induces RANK and EGFR mRNA Expression and Inhibits the EGFR Pathway at the Protein Level
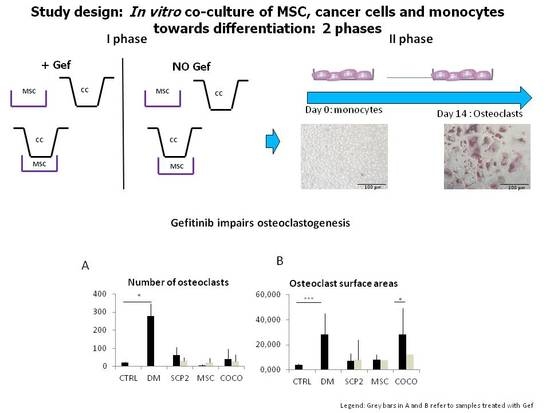
2.4. Gefitinib Impairs Osteoclastogenesis Induced by MSC-SCP2 COCO
2.5. Bone-Targeted Therapy Empower Gef Inhibition of Osteoclastogenesis
3. Discussion
4. Materials and Methods
4.1. Cell Cultures and Reagents
4.2 Drug Sensitivity Assay
4.3. MSC-SCP2 COCO Assay and Generation of CM
4.4. Generation of Pre-Osteoclasts
4.5. Osteoclastogenesis Assay with CM and Drugs
4.6. Western Blot
4.7. RNA Extraction and Real-time Quantitative PCR (qPCR)
4.8. Satistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| COCO | MSC-SCP2 co-culture |
| CM | conditioned medium |
| DM | differentiation medium |
| Gef | gefitinib |
| EGFR | epidermal growth factor receptor |
| Eve | everolimus |
| MSC | mesenchymal stromal cells |
| GF | growth factors |
| M-CSF | macrophage colony-stimulating factor |
| MTT | (4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide |
| NSCLC | non-small cell lung cancer |
| PBMC | human peripheral blood mononuclear cells |
| Den | denosumab |
| RANKL | receptor activator of nuclear factor kB ligand |
| TKI | tyrosine kinase inhibitor |
| TRAP | tartrate-resistant acid phosphatase |
| WB | western blot |
References
- Ibrahim, T.; Mercatali, L.; Amadori, D. A new emergency in oncology: Bone metastases in breast cancer patients (Review). Oncol. Lett. 2013, 6, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Coleman, R.E.; Rubens, R.D. The clinical course of bone metastases from breast cancer. Br. J. Cancer 1987, 55, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Liverani, C.; Mercatali, L.; Spadazzi, C.; La Manna, F.; De Vita, A.; Riva, N.; Calpona, S.; Ricci, M.; Bongiovanni, A.; Gunelli, E.; et al. CSF-1 blockade impairs breast cancer osteoclastogenic potential in co-culture systems. Bone 2014, 66, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Stickeler, E.; Fehm, T. Targeted and osteo-oncologic treatment in early breast cancer: What is state-of-the-art and what might become so within the next 5 years? Breast Care 2014, 9, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Weilbaecher, K.N.; Guise, T.A.; McCauley, L.K. Cancer to bone: A fatal attraction. Nat. Rev. Cancer 2011, 11, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Phinney, D.G.; Sensebe, L. Mesenchymal stromal cells: Misconceptions and evolving concepts. Cytotherapy 2013, 15, 140–145. [Google Scholar] [CrossRef] [PubMed]
- Anthony, B.A.; Link, D.C. Regulation of hematopoietic stem cells by bone marrow stromal cells. Trends Immunol. 2014, 35, 32–37. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Kang, Y. Epidermal growth factor signalling and bone metastasis. Br. J. Cancer 2010, 102, 457–461. [Google Scholar] [CrossRef] [PubMed]
- Coleman, R.E.; McCloskey, E.V. Bisphosphonates in oncology. Bone 2011, 49, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Gnant, M.; Mlineritsch, B.; Stoeger, H.; Luschin-Ebengreuth, G.; Heck, D.; Menzel, C.; Jakesz, R.; Seifert, M.; Hubalek, M.; Pristauz, G.; et al. Adjuvant endocrine therapy plus zoledronic acid in premenopausal women with early-stage breast cancer: 62-Month follow-up from the ABCSG-12 randomised trial. Lancet Oncol. 2011, 12, 631–641. [Google Scholar] [CrossRef]
- Stopeck, A.T.; Lipton, A.; Body, J.J.; Steger, G.G.; Tonkin, K.; de Boer, R.H.; Lichinitser, M.; Fujiwara, Y.; Yardley, D.A.; Viniegra, M.; et al. Denosumab compared with zoledronic acid for the treatment of bone metastases in patients with advanced breast cancer: A randomized, double-blind study. J. Clin. Oncol. 2010, 28, 5132–5139. [Google Scholar] [CrossRef] [PubMed]
- Mercatali, L.; Spadazzi, C.; Miserocchi, G.; Liverani, C.; De Vita, A.; Bongiovanni, A.; Recine, F.; Amadori, D.; Ibrahim, T. The effect of everolimus in an in vitro model of triple negative breast cancer and osteoclasts. Int. J. Mol. Sci. 2016, 17, 1827. [Google Scholar] [CrossRef] [PubMed]
- Baselga, J.; Campone, M.; Piccart, M.; Rugo, H.S.; Sahmoud, T.; Noguchi, S.; Gnant, M.; Pritchard, K.I.; Lebrun, F.; Beck, J.; et al. Eve in postmenopausal hormone-receptor-positive advanced breast cancer. N. Engl. J. Med. 2012, 366, 520–529. [Google Scholar] [CrossRef] [PubMed]
- Gnant, M.; Baselga, J.; Rugo, H.S.; Noguchi, S.; Burris, H.A.; Piccart, M.; Hortobagyi, G.N.; Eakle, J.; Mukai, H.; Iwata, H.; et al. Effect of eve on bone marker levels and progressive disease in bone in BOLERO-2. J. Natl. Cancer Inst. 2013, 105, 654–663. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R., Jr. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol. Res. 2014, 79, 34–74. [Google Scholar] [CrossRef] [PubMed]
- Foley, J.; Nickerson, N.K.; Nam, S.; Allen, K.T.; Gilmore, J.L.; Nephew, K.P.; Riese, D.J. EGFR signaling in breast cancer: Bad to the bone. Semin. Cell Dev. Biol. 2010, 21, 951–960. [Google Scholar] [CrossRef] [PubMed]
- Normanno, N.; Gullick, W.J. Epidermal growth factor receptor tyrosine kinase inhibitors and bone metastases: Different mechanisms of action for a novel therapeutic application? Endocr. Relat. Cancer 2006, 13, 3–6. [Google Scholar] [CrossRef] [PubMed]
- National Comprehensive Care Network. NCCN Clinical Practice Guidelines in Oncology: Non-Small Cell Lung Cancer V4. 2016. Available online: https://www.nccn.org/ (accessed on 15 August 2016).
- Lu, X.; Wang, Q.; Hu, G.; Van Poznak, C.; Fleisher, M.; Reiss, M.; Massagué, J.; Kang, Y. ADAMTS1 and MMP1 proteolytically engage EGF-like ligands in an osteolytic signaling cascade for bone metastasis. Genes Dev. 2009, 23, 1882–1894. [Google Scholar] [CrossRef] [PubMed]
- Ciardiello, F.; Caputo, R.; Bianco, R.; Damiano, V.; Fontanini, G.; Cuccato, S.; De Placido, S.; Bianco, A.R.; Tortora, G. Inhibition of growth factor production and angiogenesis in human cancer cells by ZD1839 (Iressa), a selective epidermal growth factor receptor tyrosine kinase inhibitor. Clin. Cancer Res. 2001, 7, 1459–1465. [Google Scholar] [PubMed]
- Moasser, M.M.; Basso, A.; Averbuch, S.D.; Rosen, N. The tyrosine kinase inhibitor ZD1839 (“Iressa”) inhibits HER2-driven signaling and suppresses the growth of ER2-overexpressing tumor cells. Cancer Res. 2001, 61, 7184–7188. [Google Scholar] [PubMed]
- Borghese, C.; Cattaruzza, L.; Pivetta, E.; Normanno, N.; de Luca, A.; Mazzucato, M.; Celegato, M.; Colombatti, A.; Aldinucci, D. Gefitinib inhibits the cross-talk between mesenchymal stem cells and prostate cancer cells leading to tumor cell proliferation and inhibition of docetaxel activity. J. Cell. Biochem. 2013, 114, 1135–1144. [Google Scholar] [CrossRef] [PubMed]
- Baselga, J.; Albanell, J.; Ruiz, A.; Lluch, A.; Gascón, P.; Guillém, V.; González, S.; Sauleda, S.; Marimón, I.; Tabernero, J.M.; et al. Phase II and tumor pharmacodynamic study of gefitinib in patients with advanced breast cancer. J. Clin. Oncol. 2005, 23, 5323–5333. [Google Scholar] [CrossRef] [PubMed]
- Lorusso, P.M. Phase I studies of ZD1839 in patients with common solid tumors. Semin. Oncol. 2003, 30, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Joensuu, G. A phase II trial of gefitinib in patients with rising PSA following radical prostatectomy or radiotherapy. Acta Oncol. 2012, 51, 130–133. [Google Scholar] [CrossRef] [PubMed]
- Osborne, C.K.; Neven, P.; Dirix, L.Y.; Mackey, J.R.; Robert, J.; Underhill, C.; Schiff, R.; Gutierrez, C.; Migliaccio, I.; Anagnostou, V.K.; et al. Gefitinib or placebo in combination with tamoxifen in patients with hormone receptor-positive metastatic breast cancer: A randomized phase II study. Clin. Cancer Res. 2011, 17, 1147–1159. [Google Scholar] [CrossRef] [PubMed]
- Somlo, G.; Martel, C.L.; Lau, S.K.; Frankel, P.; Ruel, C.; Gu, L.; Hurria, A.; Chung, C.; Luu, T.; Morgan, R., Jr.; et al. A phase I/II prospective, single arm trial of gefitinib, trastuzumab, and docetaxel in patients with stage IV HER-2 positive metastatic breast cancer. Breast Cancer Res. Treat. 2012, 131, 899–906. [Google Scholar] [CrossRef] [PubMed]
- Zampa, G.; Moscato, M.; Brannigan, B.W.; Morabito, A.; Bell, D.W.; Normanno, N. Prolonged control of bone metastases in non-small-cell lung cancer patients treated with gefitinib. Lung Cancer 2008, 60, 452–454. [Google Scholar] [CrossRef] [PubMed]
- Zukawa, M.; Nakano, M.; Hirano, N.; Mizuhashi, K.; Kanamori, M. The effectiveness of gefitinib on spinal metastases of lung cancer—Report of two cases. Asian Spine J. 2008, 2, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Von Minckwitz, G.; Jonat, W.; Fasching, P.; du Bois, A.; Kleeberg, U.; Lück, H.J.; Kettner, E.; Hilfrich, J.; Eiermann, W.; Torode, J.; et al. A multicentre phase II study on gefitinib in taxane- and anthracycline-pretreated metastatic breast cancer. Breast Cancer Res. Treat. 2005, 89, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Albain, K.S.; Gradishar, W.J.; Hayes, D.F.; Rowinsky, E.; Hudis, C.; Pusztai, L.; Tripathy, D.; Modi, S.; Rubi, S. Open-label, phase II, multicenter trial of ZD1839 (“Iressa”) in patients with advanced breast cancer. Breast Cancer Res. Treat. 2002, 76, S33. [Google Scholar]
- Ibrahim, T.; Flamini, E.; Mercatali, L.; Sacanna, E.; Serra, P.; Amadori, D. Pathogenesis of osteoblastic bone metastases from prostate cancer. Cancer 2010, 116, 1406–1418. [Google Scholar] [CrossRef] [PubMed]
- Awolaran, O.; Brooks, S.A.; Lavender, V. Breast cancer osteomimicry and its role in bone specific metastasis; an integrative, systematic review of preclinical evidence. Breast 2016, 30, 156–171. [Google Scholar] [CrossRef] [PubMed]
- Plotkin, L.; Speacht, T.L.; Donahue, H.J. Cx43 and mechanotransduction in bone. Curr. Osteoporos. Rep. 2015, 13, 67–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, D.H.; Nakashima, T.; Sanchez, O.H.; Kozieradzki, I.; Komarova, S.V.; Sarosi, I.; Morony, S.; Rubin, E.; Sarao, R.; Hojilla, C.V.; et al. Regulation of cancer cell migration and bone metastasis by RANKL. Nature 2006, 440, 692–696. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, T.; Sacanna, E.; Gaudio, M.; Mercatali, L.; Scarpi, E.; Zoli, W.; Serra, P.; Ricci, R.; Serra, L.; Kang, Y.; et al. Role of RANK, RANKL, OPG, and CXCR4 tissue markers in predicting bone metastases in breast cancer patients. Clin. Breast Cancer 2011, 11, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Smid, M.; Wang, Y.; Klijn, J.G.; Sieuwerts, A.M.; Zhang, Y.; Atkins, D.; Martens, J.W.; Foekens, J.A. Genes associated with breast cancer metastatic to bone. J. Clin. Oncol. 2006, 24, 2261–2267. [Google Scholar] [CrossRef] [PubMed]
- Bougen, N.M.; Amiry, N.; Yuan, Y.; Kong, X.J.; Pandey, V.; Vidal, L.J.; Perry, J.K.; Zhu, T.; Lobie, P.E. Trefoil factor 1 suppression of E-CADHERIN enhances prostate carcinoma cell invasiveness and metastasis. Cancer Lett. 2013, 332, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Markicevic, M.; Džodić, R.; Buta, M.; Kanjer, K.; Mandušić, V.; Nešković-Konstantinović, Z.; Nikolić-Vukosavljević, D. Trefoil factor 1 in early breast carcinoma: A potential indicator of clinical outcome during the first 3 years of follow-up. Int. J. Med. Sci. 2014, 11, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Santini, D.; Schiavon, G.; Vincenzi, B.; Gaeta, L.; Pantano, F.; Russo, A.; Ortega, C.; Porta, C.; Galluzzo, S.; Armento, G.; et al. Receptor activator of NF-κB (RANK) expression in primary tumors associates with bone metastasis occurrence in breast cancer patients. PLoS ONE 2011, 6, e19234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blake, M.L.; Tometsko, M.; Miller, S.; Jones, J.C.; Dougall, W.C. RANK expression on breast cancer cells promotes skeletal metastasis. Clin. Exp. Metastasis 2014, 31, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Kawaida, R.; Ohtsuka, T.; Okutsu, J.; Takahashi, T.; Kadono, Y.; Oda, H.; Hikita, A.; Nakamura, K.; Tanaka, S.; Furukawa, H. Jun dimerization protein 2 (JDP2), a member of the AP-1 family of transcription factor, mediates osteoclast differentiation induced by RANKL. J. Exp. Med. 2003, 197, 1029–1035. [Google Scholar] [CrossRef] [PubMed]
- Minn, A.J.; Kang, Y.; Serganova, I.; Gupta, G.P.; Giri, D.D.; Doubrovin, M.; Ponomarev, V.; Gerald, W.L.; Blasberg, R.; Massagué, J. Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J. Clin. Investig. 2005, 115, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Prockop, D.J.; Gregory, C.A.; Spees, J.L. One strategy for cell and gene therapy: Harnessing the power of adult stem cells to repair tissues. Proc. Natl. Acad. Sci. USA 2003, 100, 11917–11923. [Google Scholar] [CrossRef] [PubMed]
- Normanno, N.; de Luca, A.; Aldinucci, D.; Maiello, M.R.; Mancino, M.; D’Antonio, A.; de Filippi, R.; Pinto, A. Gefitinib inhibits the ability of human bone marrow stromal cells to induce osteoclast differentiation: Implications for the pathogenesis and treatment of bone metastasis. Endocr. Relat. Cancer 2005, 12, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, P.; Gilkes, D.M.; Wong, C.C.; Kshitiz, L.; Luo, W.; Zhang, H.; Wei, H.; Takano, N.; Schito, L.; Levchenko, A.; Semenza, G.L. Hypoxia-inducible factor-dependent breast cancer-mesenchymal stem cell bidirect sharing mediumional signaling promotes metastasis. J. Clin. Investig. 2013, 123, 189–205. [Google Scholar] [CrossRef] [PubMed]
- Karnoub, A.E.; Dash, A.B.; Vo, A.P.; Sullivan, A.; Brooks, M.W.; Bell, G.W.; Richardson, A.L.; Polyak, K.; Tubo, R.; Weinberg, R.A. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature 2007, 449, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Barcellos-de-Souza, P.; Gori, V.; Bambi, F.; Chiarugi, P. Tumor microenvironment: Bone marrow-mesenchymal stem cells as key players. Biochim. Biophys. Acta 2013, 1836, 321–335. [Google Scholar] [CrossRef] [PubMed]
- Normanno, N.; de Luca, A.; Maiello, M.R.; Campiglio, M.; Napolitano, M.; Mancino, M.; Carotenuto, A.; Viglietto, G.; Menard, S. The MEK/MAPK pathway is involved in the resistance of breast cancer cells to the EGFR tyrosine kinase inhibitor gefitinib. J. Cell. Physiol. 2006, 207, 420–427. [Google Scholar] [CrossRef] [PubMed]
- Conti, A.; Espina, V.; Chiechi, A.; Magagnoli, G.; Novello, C.; Pazzaglia, L.; Quattrini, I.; Picci, P.; Liotta, L.A.; Benassi, M.S. Mapping protein signal pathway interaction in sarcoma bone metastasis: Linkage between rank, metalloproteinases turnover and growth factor signaling pathways. Clin. Exp. Metastasis 2014, 31, 15–24. [Google Scholar] [CrossRef] [PubMed]






| Differentiation Medium | Pre-Osteoclast Medium | Pre-Osteoclast Medium | ||
|---|---|---|---|---|
| supplemented with Early-CM from: | supplemented with Late-CM from: | supplemented with Gef-CM from: | ||
| Alone (positive control) | Alone (negative control) | MSC culture | MSC culture | MSC culture |
| SCP2 culture | SCP2 culture | SCP2 culture | ||
| COCO | COCO | COCOC | ||
| SYBR™ Green Assays | |||
| Gene symbol | Forward primer (5’–3’) | Reverse primer (5’–3’) | |
| ACTB | GCACAGAGCCTCGCCTT | CCTTGCACATGCCGGAG | |
| HPRT1 | AGACTTTGCTTTCCTTGGTCAGG | GTCTGGCTTATATCCAACACTTCG | |
| SPP1 | AGATGGGTCAGGGTTTAGCC | CATCACCTGTGCCATACCAG | |
| GJA1 (CX43) | TCTGAGTGCCTGAACTTGC | ACTGACAGCCACACCTTCC | |
| ANGPT1 | CCGACTTCATGTTTTCCACA | ACCGGATTTCTCTTCCCAGA | |
| JDP2 | CTTCTTCTTGTTCCGGCATC | CTTCCTGGAGGTGAAACTGG | |
| CTSK | GCCAGACAACAGATTTCCATC | CAGAGCAAAGCTCACCAGAG | |
| TaqMan® Assays | |||
| Gene symbol | Assay identification number | ||
| ACTB | Hs99999903_m1 | ||
| HPRT1 | Hs02800695_m1 | ||
| EGFR | Hs01076078_m1 | ||
| TNFRSF11A (RANK) | Hs00921372_m1 | ||
| TFF1 | Hs00907239_m1 | ||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mercatali, L.; La Manna, F.; Miserocchi, G.; Liverani, C.; De Vita, A.; Spadazzi, C.; Bongiovanni, A.; Recine, F.; Amadori, D.; Ghetti, M.; et al. Tumor-Stroma Crosstalk in Bone Tissue: The Osteoclastogenic Potential of a Breast Cancer Cell Line in a Co-Culture System and the Role of EGFR Inhibition. Int. J. Mol. Sci. 2017, 18, 1655. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18081655
Mercatali L, La Manna F, Miserocchi G, Liverani C, De Vita A, Spadazzi C, Bongiovanni A, Recine F, Amadori D, Ghetti M, et al. Tumor-Stroma Crosstalk in Bone Tissue: The Osteoclastogenic Potential of a Breast Cancer Cell Line in a Co-Culture System and the Role of EGFR Inhibition. International Journal of Molecular Sciences. 2017; 18(8):1655. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18081655
Chicago/Turabian StyleMercatali, Laura, Federico La Manna, Giacomo Miserocchi, Chiara Liverani, Alessandro De Vita, Chiara Spadazzi, Alberto Bongiovanni, Federica Recine, Dino Amadori, Martina Ghetti, and et al. 2017. "Tumor-Stroma Crosstalk in Bone Tissue: The Osteoclastogenic Potential of a Breast Cancer Cell Line in a Co-Culture System and the Role of EGFR Inhibition" International Journal of Molecular Sciences 18, no. 8: 1655. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18081655