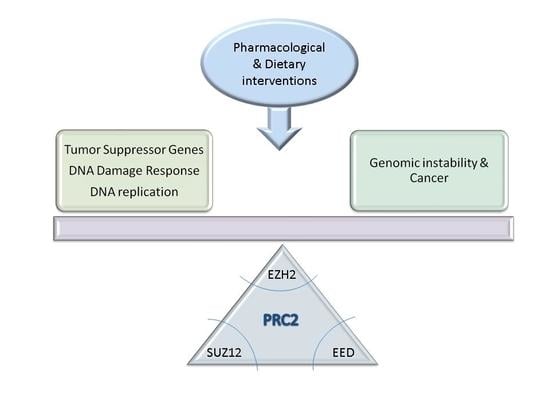
Polycomb Repressor Complex 2 in Genomic Instability and Cancer
Abstract
:1. Introduction
2. Polycomb Repressor Complex 2 Is Deregulated in Malignancy
3. Mechanisms by Which PRC2 Influences Malignant Transformation and Growth
3.1. Regulation of Tumor Suppressor Gene Expression by PRC2
3.2. Regulation of DNA Damage Response (DDR) by PRC2
3.3. PRC2 Impacts on the Fidelity of DNA Replication
4. Pharmacological and Dietary Interventions Targeting EZH2 Deregulation in Cancer
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Lewis, E.B. A gene complex controlling segmentation in Drosophila. Nature 1978, 276, 565–570. [Google Scholar] [CrossRef] [PubMed]
- Sauvageau, M.; Sauvageau, G. Polycomb Group Proteins: Multi-Faceted Regulators of Somatic Stem Cells and Cancer. Cell Stem Cell 2010, 7, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Di Croce, L.; Helin, K. Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 2013, 20, 1147–1155. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Qin, J.-J.; Voruganti, S.; Nag, S.; Zhou, J.; Zhang, R. Polycomb Group (PcG) Proteins and Human Cancers: Multifaceted Functions and Therapeutic Implications. Med. Res. Rev. 2015, 35, 1220–1267. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, D.; Imaizumi, Y.; Hasegawa, H.; Osaka, A.; Tsukasaki, K.; Choi, Y.L.; Mano, H.; Marquez, V.E.; Hayashi, T.; Yanagihara, K.; Moriwaki, Y. Overexpression of Enhancer of zeste homolog 2 with trimethylation of lysine 27 on histone H3 in adult T-cell leukemia/lymphoma as a target for epigenetic therapy. Haematologica 2011, 96, 712–719. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Hu, C.; Arnovitz, S.; Bugno, J.; Yu, M.; Zuo, Z.; Chen, P.; Huang, H.; Ulrich, B.; Gurbuxani, S.; Weng, H. miR-22 has a potent anti-tumour role with therapeutic potential in acute myeloid leukaemia. Nat. Commun. 2016, 7, 11452. [Google Scholar] [CrossRef] [PubMed]
- Toll, A.D.; Dasgupta, A.; Potoczek, M.; Yeo, C.J.; Kleer, C.G.; Brody, J.R.; Witkiewicz, A.K. Implications of enhancer of zeste homologue 2 expression in pancreatic ductal adenocarcinoma. Hum. Pathol. 2010, 41, 1205–1209. [Google Scholar] [CrossRef] [PubMed]
- Han, T.; Jiao, F.; Hu, H.; Yuan, C.; Wang, L.; Jin, Z.L.; Song, W.F.; Wang, L.W. EZH2 promotes cell migration and invasion but not alters cell proliferation by suppressing E-cadherin, partly through association with MALAT-1 in pancreatic cancer. Oncotarget 2016, 7, 11194–11207. [Google Scholar] [CrossRef] [PubMed]
- Avan, A.; Crea, F.; Paolicchi, E.; Funel, N.; Galvani, E.; Marquez, V.E.; Honeywell, R.J.; Danesi, R.; Peters, G.J.; Giovannetti, E. Molecular mechanisms involved in the synergistic interaction of the EZH2 inhibitor 3-deazaneplanocin A with gemcitabine in pancreatic cancer cells. Mol. Cancer Ther. 2012, 11, 1735–1746. [Google Scholar] [CrossRef] [PubMed]
- Mallen-St Clair, J.; Soydaner-Azeloglu, R.; Lee, K.E.; Taylor, L.; Livanos, A.; Pylayeva-Gupta, Y.; Miller, G.; Margueron, R.; Reinberg, D.; Bar-Sagi, D. EZH2 couples pancreatic regeneration to neoplastic progression. Genes. Dev. 2012, 26, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Varambally, S.; Dhanasekaran, S.M.; Zhou, M.; Barrette, T.R. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002, 419, 624–629. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Deng, Z.; Wan, M.; Huang, W.; Cramer, S.D.; Xu, J.; Lei, M.; Sui, G. MicroRNA-101 negatively regulates Ezh2 and its expression is modulated by androgen receptor and HIF-1α/HIF-1β. Mol. Cancer 2010, 9, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleer, C.G.; Cao, Q.; Varambally, S.; Shen, R.; Ota, I.; Tomlins, S.A.; Ghosh, D.; Sewalt, R.G.; Otte, A.P.; Hayes, D.F.; Sabel, M.S. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. USA 2003, 100, 11606–11611. [Google Scholar] [CrossRef] [PubMed]
- Zeidler, M.; Varambally, S.; Cao, Q.; Chinnaiyan, A.M.; Ferguson, D.O.; Merajver, S.D.; Kleer, C.G. The Polycomb group protein EZH2 impairs DNA repair in breast epithelial cells. Neoplasia 2005, 7, 1011–1019. [Google Scholar] [CrossRef] [PubMed]
- Raman, J.D.; Mongan, N.P.; Tickoo, S.K.; Boorjian, S.A.; Scherr, D.S.; Gudas, L.J. Increased expression of the polycomb group gene, EZH2, in transitional cell carcinoma of the bladder. Clin. Cancer Res. 2005, 11, 8570–8576. [Google Scholar] [CrossRef] [PubMed]
- Arisan, S.; Buyuktuncer, E.D.; Palavan-Unsal, N.; Caskurlu, T.; Cakir, O.O.; Ergenekon, E. Increased expression of EZH2, a polycomb group protein, in bladder carcinoma. Urol. Int. 2005, 75, 252–257. [Google Scholar] [CrossRef] [PubMed]
- Weikert, S.; Christoph, F.; Kollermann, J.; Muller, M.; Schrader, M.; Miller, K.; Schrader, M.; Miller, K.; Krause, H. Expression levels of the EZH2 polycomb transcriptional repressor correlate with aggressiveness and invasive potential of bladder carcinomas. Int. J. Mol. Med. 2005, 16, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.H.; Huang, H.S.; Wu, H.U.; Tsai, Y.T.; Chuang, M.J.; Yu, C.P.; Huang, S.M.; Sun, G.H.; Chang, S.Y.; Hsiao, P.W.; Yu, D.S. Pharmacologic down-regulation of EZH2 suppresses bladder cancer in vitro and in vivo. Oncotarget 2014, 5, 10342–10355. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xiang, W.; Wang, M.; Huang, T.; Xiao, X.; Wang, L.; Tao, D.; Dong, L.; Zeng, F.; Jiang, G. Methyl jasmonate sensitizes human bladder cancer cells to gambogic acid-induced apoptosis through down-regulation of EZH2 expression by miR-101. Br. J. Pharmacol. 2014, 171, 618–635. [Google Scholar] [CrossRef] [PubMed]
- Matsukawa, Y.; Semba, S.; Kato, H.; Ito, A.; Yanagihara, K.; Yokozaki, H. Expression of the enhancer of zeste homolog 2 is correlated with poor prognosis in human gastric cancer. Cancer Sci. 2006, 97, 484–491. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.T.; Zhu, F.; Lin, W.R.; Ying, R.B.; Yang, Y.P.; Zeng, L.H. The novel EZH2 inhibitor, GSK126, suppresses cell migration and angiogenesis via down-regulating VEGF-A. Cancer Chemother. Pharmacol. 2016, 77, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Yuan, C.; Xiao, X.; Wang, X.; Ji, X.; Yu, H.; Wu, Z.; Zhang, J. The expression and significance of the enhancer of zeste homolog 2 in lung adenocarcinoma. Oncol. Rep. 2012, 28, 147–154. [Google Scholar] [PubMed]
- Huqun; Ishikawa, R.; Zhang, J.; Miyazawa, H.; Goto, Y.; Shimizu, Y. Enhancer of zeste homolog 2 is a novel prognostic biomarker in nonsmall cell lung cancer. Cancer 2012, 118, 1599–1606. [Google Scholar]
- Frankel, A.E.; Liu, X.; Minna, J.D. Developing EZH2-Targeted Therapy for Lung Cancer. Cancer Discov. 2016, 6, 949–952. [Google Scholar] [CrossRef] [PubMed]
- Berns, K.; Berns, A. Awakening of “Schlafen11” to Tackle Chemotherapy Resistance in SCLC. Cancer Cell 2017, 31, 169–171. [Google Scholar] [CrossRef] [PubMed]
- Sudo, T.; Utsunomiya, T.; Mimori, K.; Nagahara, H.; Ogawa, K.; Inoue, H.; Wakiyama, S.; Fujita, H.; Shirouzu, K.; Mori, M. Clinicopathological significance of EZH2 mRNA expression in patients with hepatocellular carcinoma. Br. J. Cancer 2005, 92, 1754–1758. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.Y.; Tong, Z.T.; Zheng, F.; Liao, Y.J.; Wang, Y.; Rao, H.L.; Chen, Y.C.; Wu, Q.L.; Liu, Y.H.; Guan, X.Y.; Lin, M.C. EZH2 protein: A promising immunomarker for the detection of hepatocellular carcinomas in liver needle biopsies. Gut 2011, 60, 967–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasaki, M.; Ikeda, H.; Itatsu, K.; Yamaguchi, J.; Sawada, S.; Minato, H.; Ohta, T.; Nakanuma, Y. The overexpression of polycomb group proteins Bmi1 and EZH2 is associated with the progression and aggressive biological behavior of hepatocellular carcinoma. Lab Invest. 2008, 88, 873–882. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.P.; Hong, Y.H.; Tung, K.Y.; Yang, P.M. In silico and experimental analyses predict the therapeutic value of an EZH2 inhibitor GSK343 against hepatocellular carcinoma through the induction of metallothionein genes. Oncoscience 2016, 3, 9–20. [Google Scholar] [PubMed]
- Liu, T.P.; Lo, H.L.; Wei, L.S.; Hsiao, H.H.; Yang, P.M. S-Adenosyl-l-methionine-competitive inhibitors of the histone methyltransferase EZH2 induce autophagy and enhance drug sensitivity in cancer cells. Anticancer Drugs 2015, 26, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Ahani, N.; Shirkoohi, R.; Rokouei, M.; Alipour Eskandani, M.; Nikravesh, A. Overexpression of enhancer of zeste human homolog 2 (EZH2) gene in human cytomegalovirus positive glioblastoma multiforme tissues. Med. Oncol. 2014, 31, 252. [Google Scholar] [CrossRef] [PubMed]
- Suva, M.L.; Riggi, N.; Janiszewska, M.; Radovanovic, I.; Provero, P.; Stehle, J.C.; Baumer, K.; Le Bitoux, M.A.; Marino, D.; Cironi, L.; Marquez, V.E. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res. 2009, 69, 9211–9218. [Google Scholar] [CrossRef] [PubMed]
- Holland, D.; Hoppe-Seyler, K.; Schuller, B.; Lohrey, C.; Maroldt, J.; Durst, M.; Hoppe-Seyler, F. Activation of the enhancer of zeste homologue 2 gene by the human papillomavirus E7 oncoprotein. Cancer Res. 2008, 68, 9964–9972. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Yang, Z.; Ye, W.; Yu, X.; Hua, X. Prognostic significance of histone methyltransferase enhancer of zeste homolog 2 in patients with cervical squamous cell carcinoma. Oncol. Lett. 2015, 10, 857–862. [Google Scholar] [CrossRef] [PubMed]
- Rao, Z.Y.; Cai, M.Y.; Yang, G.F.; He, L.R.; Mai, S.J.; Hua, W.F.; Liao, Y.J.; Deng, H.X.; Chen, Y.C.; Guan, X.Y.; Zeng, Y.X. EZH2 supports ovarian carcinoma cell invasion and/or metastasis via regulation of TGF-β1 and is a predictor of outcome in ovarian carcinoma patients. Carcinogenesis 2010, 31, 1576–1583. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, I.M.; Halvorsen, O.J.; Collett, K.; Stefansson, I.M.; Straume, O.; Haukaas, S.A.; Salvesen, H.B.; Otte, A.P.; Akslen, L.A. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J. Clin. Oncol. 2006, 24, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Tiffen, J.C.; Gunatilake, D.; Gallagher, S.J.; Gowrishankar, K.; Heinemann, A.; Cullinane, C.; Dutton-Regester, K.; Pupo, G.M.; Strbenac, D.; Yang, J.Y.; Madore, J. Targeting activating mutations of EZH2 leads to potent cell growth inhibition in human melanoma by derepression of tumor suppressor genes. Oncotarget 2015, 6, 27023–27036. [Google Scholar] [CrossRef] [PubMed]
- Zingg, D.; Debbache, J.; Schaefer, S.M.; Tuncer, E.; Frommel, S.C.; Cheng, P.; Arenas-Ramirez, N.; Haeusel, J.; Zhang, Y.; Bonalli, M.; McCabe, M.T. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat. Commun. 2015, 6, 6051. [Google Scholar] [CrossRef] [PubMed]
- Changchien, Y.C.; Tatrai, P.; Papp, G.; Sapi, J.; Fonyad, L.; Szendroi, M.; Pápai, Z.; Sápi, Z. Poorly differentiated synovial sarcoma is associated with high expression of enhancer of zeste homologue 2 (EZH2). J. Transl. Med. 2012, 10, 216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciarapica, R.; Miele, L.; Giordano, A.; Locatelli, F.; Rota, R. Enhancer of zeste homolog 2 (EZH2) in pediatric soft tissue sarcomas: first implications. BMC Med. 2011, 9, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Visser, H.P.; Gunster, M.J.; Kluin-Nelemans, H.C.; Manders, E.M.; Raaphorst, F.M.; Meijer, C.J.; Willemze, R.; Otte, A.P. The Polycomb group protein EZH2 is upregulated in proliferating, cultured human mantle cell lymphoma. Br. J. Haematol. 2001, 112, 950–958. [Google Scholar] [CrossRef] [PubMed]
- Kodach, L.L.; Jacobs, R.J.; Heijmans, J.; van Noesel, C.J.; Langers, A.M.; Verspaget, H.W.; Hommes, D.W.; Offerhaus, G.J.A.; van den Brink, G.R.; Hardwick, J.C. The role of EZH2 and DNA methylation in the silencing of the tumour suppressor RUNX3 in colorectal cancer. Carcinogenesis 2010, 31, 1567–1575. [Google Scholar] [CrossRef] [PubMed]
- Fluge, O.; Gravdal, K.; Carlsen, E.; Vonen, B.; Kjellevold, K.; Refsum, S.; Lilleng, R.; Eide, T.J.; Halvorsen, T.B.; Tveit, K.M.; Otte, A.P. Expression of EZH2 and Ki-67 in colorectal cancer and associations with treatment response and prognosis. Br. J. Cancer 2009, 101, 1282–1289. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.G.; Ye, Y.J.; Yuan, J.; Liu, F.F.; Zhang, H.; Wang, S. EZH2 and STAT6 expression profiles are correlated with colorectal cancer stage and prognosis. World J. Gastroenterol. 2010, 16, 2421–2427. [Google Scholar] [CrossRef] [PubMed]
- Lei, Q.; Shen, F.; Wu, J.; Zhang, W.; Wang, J.; Zhang, L. MiR-101, downregulated in retinoblastoma, functions as a tumor suppressor in human retinoblastoma cells by targeting EZH2. Oncol. Rep. 2014, 32, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, X.; Chen, Z.; Huang, H.; Jin, Y.; Kolokythas, A.; Wang, A.; Dai, Y.; Wong, D.T.; Zhou, X. Polycomb group protein EZH2-mediated E-cadherin repression promotes metastasis of oral tongue squamous cell carcinoma. Mol. Carcinog. 2013, 52, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, Y.; Qiu, J.; Li, Q.; Yuan, C.; Zhang, W.; Wang, D.; Ye, J.; Jiang, H.; Yang, J.; Cheng, J. The polycomb group protein EZH2 is a novel therapeutic target in tongue cancer. Oncotarget 2013, 4, 2532–2549. [Google Scholar] [CrossRef] [PubMed]
- De Raedt, T.; Beert, E.; Pasmant, E.; Luscan, A.; Brems, H.; Ortonne, N.; Helin, K.; Hornick, J.L.; Mautner, V.; Kehrer-Sawatzki, H.; Clapp, W. PRC2 loss amplifies Ras-driven transcription and confers sensitivity to BRD4-based therapies. Nature 2014, 514, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Vainchenker, W.; Delhommeau, F.; Constantinescu, S.N.; Bernardm, O.A. New mutations and pathogenesis of myeloproliferative neoplasms. Blood 2011, 118, 1723–1735. [Google Scholar] [CrossRef] [PubMed]
- Nikoloski, G.; Langemeijer, S.M.; Kuiper, R.P.; Knops, R.; Massop, M.; Tonnissen, E.R.; Van der Heijden, A.; Scheele, T.N.; Vandenberghe, P.; de Witte, T.; et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat. Genet. 2010, 42, 665–667. [Google Scholar] [CrossRef] [PubMed]
- Ernst, T.; Chase, A.J.; Score, J.; Hidalgo-Curtis, C.E.; Bryant, C.; Jones, A.V.; Waghorn, K.; Zoi, K.; Ross, F.M.; Reiter, A.; Hochhaus, A. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat. Genet. 2010, 42, 722–726. [Google Scholar] [CrossRef] [PubMed]
- Venneti, S.; Le, P.; Martinez, D.; Xie, S.X.; Sullivan, L.M.; Rorke-Adams, L.B.; Pawel, B.; Judkins, A.R. Malignant rhabdoid tumors express stem cell factors, which relate to the expression of EZH2 and Id proteins. Am. J. Surg. Pathol. 2011, 35, 1463–1472. [Google Scholar] [CrossRef] [PubMed]
- Ntziachristos, P.; Tsirigos, A.; Van Vlierberghe, P.; Nedjic, J.; Trimarchi, T.; Flaherty, M.S.; Ferres-Marco, D.; Da Ros, V.; Tang, Z.; Siegle, J.; Asp, P. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat. Med. 2012, 18, 298–301. [Google Scholar] [CrossRef] [PubMed]
- Morin, R.D.; Johnson, N.A.; Severson, T.M.; et al. Somatic mutation of EZH2 (Y641) in Follicular and Diffuse Large B-cell Lymphomas of Germinal Center Origin. Nat. Genet. 2010, 42, 181–185. [Google Scholar] [CrossRef] [PubMed]
- McCabe, M.T.; Graves, A.P.; Ganji, G.; Diaz, E.; Halsey, W.S.; Jiang, Y.; Smitheman, K.N.; Ott, H.M.; Pappalardi, M.B.; Allen, K.E.; Chen, S.B. Mutation of A677 in histone methyltransferase EZH2 in human B-cell lymphoma promotes hypertrimethylation of histone H3 on lysine 27 (H3K27). Proc. Natl. Acad. Sci. USA 2012, 109, 2989–2994. [Google Scholar] [CrossRef] [PubMed]
- Majer, C.R.; Jin, L.; Scott, M.P.; Knutson, S.K.; Kuntz, K.W.; Keilhack, H.; Smith, J.J.; Moyer, M.P.; Richon, V.M.; Copeland, R.A.; Wigle, T.J. A687V EZH2 is a gain-of-function mutation found in lymphoma patients. FEBS Lett. 2012, 586, 3448–3451. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Chan, H.; Teng, L.; Li, L.; Chuai, S.; Zhang, R.; Zeng, J.; Li, M.; Fan, H.; Lin, Y.; Gu, J. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc. Natl. Acad. Sci. USA 2012, 109, 21360–21365. [Google Scholar] [CrossRef] [PubMed]
- Knutson, S.K.; Wigle, T.J.; Warholic, N.M.; Sneeringer, C.J.; Allain, C.J.; Klaus, C.R.; et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat. Chem. Biol. 2012, 8, 890–896. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Simons, D.L.; Segall, I.; Carcamo-Cavazos, V.; Schwartz, E.J.; Yan, N.; Zuckerman, N.S.; Dirbas, F.M.; Johnson, D.L.; Holmes, S.P.; Lee, P.P. PRC2/EED-EZH2 complex is up-regulated in breast cancer lymph node metastasis compared to primary tumor and correlates with tumor proliferation in situ. PLoS ONE 2012, 7, e51239. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.L.; Gao, X.; Jiang, Y.; Zhang, G.; Sun, Z.C.; Cui, B.B.; Yang, Y.M. Expression and clinicopathological significance of EED, SUZ12 and EZH2 mRNA in colorectal cancer. J. Cancer Res. Clin. Oncol. 2015, 141, 661–669. [Google Scholar] [CrossRef] [PubMed]
- Kirmizis, A.; Bartley, S.M.; Farnham, P.J. Identification of the polycomb group protein SU(Z)12 as a potential molecular target for human cancer therapy. Mol. Cancer Ther. 2003, 2, 113–121. [Google Scholar] [PubMed]
- Chen, H.; Gao, S.; Li, J.; Liu, D.; Sheng, C.; Yao, C.; Jiang, W.; Wu, J.; Chen, S.; Huang, W. Wedelolactone disrupts the interaction of EZH2-EED complex and inhibits PRC2-dependent cancer. Oncotarget 2015, 6, 13049–13059. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Fillmore Brainson, C.; Koyama, S.; Redig, A.J.; Chen, T.; Li, S.; Gupta, M.; Garcia-de-Alba, C.; Paschini, M.; Herter-Sprie, G.S.; Lu, G. Lkb1 inactivation drives lung cancer lineage switching governed by Polycomb Repressive Complex 2. Nat. Commun. 2017, 8, 14922. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Teckie, S.; Wiesner, T.; Ran, L.; Prieto Granada, C.N.; Lin, M.; Zhu, S.; Cao, Z.; Liang, Y.; Sboner, A.; Tap, W.D. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat. Genet. 2014, 46, 1227–1232. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Cai, Q.; Wu, H.; Vathipadiekal, V.; Dobbin, Z.C.; Li, T.; Hua, X.; Landen, C.N.; Birrer, M.J.; Sánchez-Beato, M.; Zhang, R. SUZ12 promotes human epithelial ovarian cancer by suppressing apoptosis via silencing HRK. Mol. Cancer Res. 2012, 10, 1462–1472. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Chen, J.; He, Z.; Xiao, Y. SUZ12 depletion suppresses the proliferation of gastric cancer cells. Cell. Physiol. Biochem. 2013, 31, 778–784. [Google Scholar] [CrossRef] [PubMed]
- Xia, R.; Jin, F.Y.; Lu, K.; Wan, L.; Xie, M.; Xu, T.P.; De, W.; Wang, Z.X. SUZ12 promotes gastric cancer cell proliferation and metastasis by regulating KLF2 and E-cadherin. Tumour Biol. 2015, 36, 5341–5351. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Shi, X.; Wang, L.; Wu, Y.; Jin, F.; Bai, C.; Song, Y. SUZ12 is involved in progression of non-small cell lung cancer by promoting cell proliferation and metastasis. Tumour Biol. 2014, 35, 6073–6082. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.R.; Roh, Y.G.; Kim, S.K.; Lee, J.S.; Seol, S.Y.; Lee, H.H.; Kim, W.T.; Kim, W.J.; Heo, J.; Cha, H.J.; Kang, T.H. Activation of EZH2 and SUZ12 Regulated by E2F1 Predicts the Disease Progression and Aggressive Characteristics of Bladder Cancer. Clin. Cancer Res. 2015, 21, 5391–5403. [Google Scholar] [CrossRef] [PubMed]
- Gibson, W.T.; Hood, R.L.; Zhan, S.H.; Bulman, D.E.; Fejes, A.P.; Moore, R.; Mungall, A.J.; Eydoux, P.; Babul-Hirji, R.; An, J.; Marra, M.A. Mutations in EZH2 Cause Weaver Syndrome. Am. J. Hum. Genet. 2012, 90, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Tatton-Brown, K.; Hanks, S.; Ruark, E.; Zachariou, A.; Duarte, S.D.V.; Ramsay, E.; Snape, K.; Murray, A.; Perdeaux, E.R.; Seal, S.; Loveday, C. Germline mutations in the oncogene EZH2 cause Weaver syndrome and increased human height. Oncotarget 2011, 2, 1127–1133. [Google Scholar] [CrossRef] [PubMed]
- Imagawa, E.; Higashimoto, K.; Sakai, Y.; Numakura, C.; Okamoto, N.; Matsunaga, S.; Ryo, A.; Sato, Y.; Sanefuji, M.; Ihara, K.; Takada, Y. Mutations in genes encoding polycomb repressive complex 2 subunits cause Weaver syndrome. Hum. Mutat. 2017, 38, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Cohen, A.S.A.; Yap, D.B.; Lewis, M.E.S.; Chijiwa, C.; Ramos-Arroyo, M.A.; Tkachenko, N.; Milano, V.; Fradin, M.; McKinnon, M.L.; Townsend, K.N.; Xu, J. Weaver Syndrome-Associated EZH2 Protein Variants Show Impaired Histone Methyltransferase Function In Vitro. Hum. Mutat. 2016, 37, 301–307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coulter, D.; Powell, C.M.; Gold, S. Weaver syndrome and neuroblastoma. J. Pediatr. Hematol. Oncol. 2008, 30, 758–760. [Google Scholar] [CrossRef] [PubMed]
- Huffman, C.; McCandless, D.; Jasty, R.; Matloub, J.; Robinson, H.B.; Weaver, D.D.; Cohen, M.M. Weaver syndrome with neuroblastoma and cardiovascular anomalies. Am. J. Med. Genet. 2001, 99, 252–255. [Google Scholar] [CrossRef]
- Kehrer-Sawatzki, H.; Mautner, V.-F.; Cooper, D.N. Emerging genotype–phenotype relationships in patients with large NF1 deletions. Hum. Genet. 2017, 136, 349–376. [Google Scholar] [CrossRef] [PubMed]
- Cleven, A.H.G.; Al Sannaa, G.A.; Bruijn, I.B.; Ingram, D.R.; van de Rijn, M.; Rubin, B.P.; de Vries, M.; Watson, K.; Torres, K.E.; Wang, W.L.; van Duinen, S. Loss of H3K27 tri-methylation is a diagnostic marker for Malignant Peripheral Nerve Sheath Tumors and an indicator for an inferior survival. Mod. Pathol. 2016, 29, 582–590. [Google Scholar] [CrossRef] [PubMed]
- Issa, J.-P.J. The myelodysplastic syndrome as a prototypical epigenetic disease. Blood 2013, 121, 3811–3817. [Google Scholar] [CrossRef] [PubMed]
- Muto, T.; Sashida, G.; Oshima, M.; Mochizuki-Kashio, M.; Nagata, Y.; Sanada, M.; Miyagi, S.; Saraya, A.; Kamio, A.; Nagae, G. Concurrent loss of Ezh2 and Tet2cooperates in the pathogenesis of myelodysplastic disorders. J. Exp. Med. 2013, 210, 2627–2639. [Google Scholar] [CrossRef] [PubMed]
- Sneeringer, C.J.; Scott, M.P.; Kuntz, K.W.; Knutson, S.K.; Pollock, R.M.; Richon, V.M.; Copeland, R.A. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc. Natl. Acad. Sci. USA 2010, 107, 20980–20985. [Google Scholar] [CrossRef] [PubMed]
- Yap, D.B.; Chu, J.; Berg, T.; Schapira, M.; Cheng, S.W.G.; Moradian, A.; Morin, R.D.; Mungall, A.J.; Meissner, B.; Boyle, M.; Marquez, V.E. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood 2011, 117, 2451–2459. [Google Scholar] [CrossRef] [PubMed]
- Papakonstantinou, N.; Ntoufa, S.; Chartomatsidou, E.; Kotta, K.; Agathangelidis, A.; Giassafaki, L.; Karamanli, T.; Bele, P.; Moysiadis, T.; Baliakas, P.; et al. The histone methyltransferase EZH2 as a novel prosurvival factor in clinically aggressive chronic lymphocytic leukemia. Oncotarget 2016, 7, 35946–35959. [Google Scholar] [CrossRef] [PubMed]
- Martín-Pérez, D.; Sánchez, E.; Maestre, L.; Suela, J.; Vargiu, P.; Di Lisio, L.; Martínez, N.; Alves, J.; Piris, M.A.; Sánchez-Beato, M. Deregulated Expression of the Polycomb-Group Protein SUZ12 Target Genes Characterizes Mantle Cell Lymphoma. Am. J. Pathol. 2010, 177, 930–942. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.; Gupta, K.P. Modulation of miR-203 and its regulators as a function of time during the development of 7, 12 dimethylbenz [a] anthracene induced mouse skin tumors in presence or absence of the antitumor agents. Toxicol. Appl. Pharmacol. 2014, 278, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.-G.; Kim, D.J.; Li, S.; Lee, K.Y.; Li, X.; Bode, A.M.; Dong, Z. Polycomb (PcG) Proteins, BMI1 and SUZ12, Regulate Arsenic-induced Cell Transformation. J. Biol. Chem. 2012, 287, 31920–31928. [Google Scholar] [CrossRef] [PubMed]
- Hussain, M.; Rao, M.; Humphries, A.E.; Hong, J.A.; Liu, F.; Yang, M.; Caragacianu, D.; Schrump, D.S. Tobacco smoke induces polycomb-mediated repression of Dickkopf-1 in lung cancer cells. Cancer Res. 2009, 69, 3570–3578. [Google Scholar] [CrossRef] [PubMed]
- Cha, T.L.; Zhou, B.P.; Xia, W.; Wu, Y.; Yang, C.C.; Chen, C.T.; Ping, B.; Otte, A.P.; Hung, M.C. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 2005, 310, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Sarma, K.; Margueron, R.; Ivanov, A.; Pirrotta, V.; Reinberg, D. Ezh2 Requires PHF1 To Efficiently Catalyze H3 Lysine 27 Trimethylation In Vivo. Mol. Cell. Biol. 2008, 28, 2718–2731. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.T.; Li, Z.; Wu, Z.; Aau, M.; Guan, P.; Karuturi, R.M.; Liou, Y.C.; Yu, Q. Context-specific regulation of NFKB target gene expression by EZH2 in breast cancers. Mol. Cell 2011, 43, 798–810. [Google Scholar] [CrossRef] [PubMed]
- Yap, K.L.; Li, S.; Muñoz-Cabello, A.M.; Raguz, S.; Zeng, L.; Mujtaba, S.; Gil, J.; Walsh, M.J.; Zhou, M.M. Molecular Interplay of the Non-coding RNA ANRIL and Methylated Histone H3 Lysine 27 by Polycomb CBX7 in Transcriptional Silencing of INK4a. Mol. Cell 2010, 38, 662–674. [Google Scholar] [CrossRef] [PubMed]
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15INK4B tumor suppressor gene. Oncogene 2011, 30, 1956–1962. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Milyavsky, M.; Shats, I.; Erez, N.; Goldfinger, N.; Rotter, V. Activated p53 suppresses the histone methyltransferase EZH2 gene. Oncogene 2004, 23, 5759–5769. [Google Scholar] [CrossRef] [PubMed]
- Bracken, A.P.; Pasini, D.; Capra, M.; Prosperini, E.; Colli, E.; Helin, K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003, 22, 5323–5335. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhou, Y.; Cheng, C.; Cui, H.; Cheng, L.; Kong, P.; Wang, J.; Li, Y.; Chen, W.; Song, B.; Wang, F. Genomic Analyses Reveal Mutational Signatures and Frequently Altered Genes in Esophageal Squamous Cell Carcinoma. Am. J. Hum. Genet. 2015, 96, 597–611. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, N.; Bracken, A.P.; Trinh, E.; Schjerling, C.K.; Koseki, H.; Rappsilber, J.; Helin, K.; Hansen, K.H. Bypass of senescence by the polycomb group protein CBX8 through direct binding to the INK4A-ARF locus. EMBO J. 2007, 26, 1637–1648. [Google Scholar] [CrossRef] [PubMed]
- Beguelin, W.; Teater, M.; Gearhart, M.D.; Calvo Fernandez, M.T.; Goldstein, R.L.; Cardenas, M.G.; Hatzi, K.; Rosen, M.; Shen, H.; Corcoran, C.M.; et al. EZH2 and BCL6 cooperate to assemble CBX8-BCOR complex to repress bivalent promoters, mediate germinal center formation and lymphomagenesis. Cancer Cell 2016, 30, 197–213. [Google Scholar] [CrossRef] [PubMed]
- Nie, F.Q.; Sun, M.; Yang, J.S.; Xie, M.; Xu, T.P.; Xia, R.; Liu, Y.W.; Liu, X.H.; Zhang, E.B.; Lu, K.H.; Shu, Y.Q. Long noncoding RNA ANRIL promotes non-small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol. Cancer Ther. 2015, 14, 268–277. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Ochiai, A. Enhancer of zeste homolog 2 downregulates E-cadherin by mediating histone H3 methylation in gastric cancer cells. Cancer Sci. 2008, 99, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Cao, Q.; Yu, J.; Dhanasekaran, S.M.; Kim, J.H.; Mani, R.S.; Tomlins, S.A.; Mehra, R.; Laxman, B.; Cao, X.; Yu, J.; Kleer, C.G. Repression of E-cadherin by the polycomb group protein EZH2 in cancer. Oncogene 2008, 27, 7274–7284. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.M.; Abate-Shen, C. Molecular genetics of prostate cancer: New prospects for old challenges. Genes. Dev. 2010, 24, 1967–2000. [Google Scholar] [CrossRef] [PubMed]
- Bohrer, L.R.; Chen, S.; Hallstrom, T.C.; Huang, H. Androgens suppress EZH2 expression via retinoblastoma (RB) and p130-dependent pathways: A potential mechanism of androgen-refractory progression of prostate cancer. Endocrinology 2010, 151, 5136–5145. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.C.; Yu, J.; Runkle, C.; Wu, L.; Hu, M.; Wu, D.; Liu, J.S.; Wang, Q.; Qin, Z.S.; Yu, J. Cooperation between Polycomb and androgen receptor during oncogenic transformation. Genome Res. 2012, 22, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Kunderfranco, P.; Mello-Grand, M.; Cangemi, R.; Mensah, A.; Albertini, V.; Malek, A.; Chiorino, G.; Catapano, C.V.; Carbone, G.M. ETS transcription factors control transcription of EZH2 and epigenetic silencing of the tumor suppressor gene Nkx3.1 in prostate cancer. PloS ONE 2010, 5, e10547. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Yu, J.; Rhodes, D.R.; Tomlins, S.A.; Cao, X.; Chen, G.; Mehra, R.; Wang, X.; Ghosh, D.; Shah, R.B.; Varambally, S. A polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer Res. 2007, 67, 10657–10663. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Yu, J.; Mani, R.S.; Cao, Q.; Brenner, C.J.; Cao, X.; Wang, X.; Wu, L.; Li, J.; Hu, M.; Gong, Y. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell 2010, 17, 443–454. [Google Scholar] [CrossRef] [PubMed]
- Gorgoulis, V.G.; Vassiliou, L.V.; Karakaidos, P.; Zacharatos, P. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 2005, 434, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Bartkova, J.; Horejsi, Z.; Koed, K.; Kramer, A.; Tort, F.; Zieger, K.; Guldberg, P.; Sehested, M.; Nesland, J.M.; Lukas, C.; et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 2005, 434, 864–870. [Google Scholar] [CrossRef] [PubMed]
- Halazonetis, T.D.; Gorgoulis, V.G.; Bartek, J. An oncogene-induced DNA damage model for cancer development. Science 2008, 319, 1352–1355. [Google Scholar] [CrossRef] [PubMed]
- Sarni, D.; Kerem, B. Oncogene-Induced Replication Stress Drives Genome Instability and Tumorigenesis. Int. J. Mol. Sci. 2017, 18, 1339. [Google Scholar] [CrossRef]
- Chou, D.M.; Adamson, B.; Dephoure, N.E.; Tan, X.; Nottke, A.C.; Hurov, K.E.; Gygi, S.P.; Colaiácovo, M.P.; Elledge, S.J. A chromatin localization screen reveals poly (ADP ribose)-regulated recruitment of the repressive polycomb and NuRD complexes to sites of DNA damage. Proc. Natl. Acad. Sci. 2010, 107, 18475–18480. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Jiang, J.; Lan, L.; Nakajima, S.; Kanno, S.I.; Koseki, H.; Yasui, A. A polycomb group protein, PHF1, is involved in the response to DNA double-strand breaks in human cell. Nucl. Acids Res. 2008, 36, 2939–2947. [Google Scholar] [CrossRef] [PubMed]
- O’Hagan, H.M.; Mohammad, H.P.; Baylin, S.B. Double Strand Breaks Can Initiate Gene Silencing and SIRT1-Dependent Onset of DNA Methylation in an Exogenous Promoter CpG Island. PLoS Genet. 2008, 4, e1000155. [Google Scholar] [CrossRef] [PubMed]
- Seiler, D.M.; Rouquette, J.; Schmid, V.J.; Strickfaden, H.; Ottmann, C.; Drexler, G.A.; Mazurek, B.; Greubel, C.; Hable, V.; Dollinger, G.; et al. Double-strand break-induced transcriptional silencing is associated with loss of tri-methylation at H3K4. Chromosome Res. 2011, 19, 883–899. [Google Scholar] [CrossRef] [PubMed]
- Rouleau, M.; McDonald, D.; Gagné, P.; Ouellet, M.E.; Droit, A.; Hunter, J.M.; Dutertre, S.; Prigent, C.; Hendzel, M.J.; Poirier, G.G. PARP-3 associates with polycomb group bodies and with components of the DNA damage repair machinery. J. Cell. Biochem. 2007, 100, 385–401. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.; Ismail, I.H.; Young, L.C.; Poirier, G.G.; Hendzel, M.J. Polycomb repressive complex 2 contributes to DNA double-strand break repair. Cell. Cycle 2013, 12, 2675–2683. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Lee, S.T.; Qiao, Y.; Li, Z.; Lee, P.L.; Lee, Y.J.; Jiang, X.; Tan, J.; Aau, M.; Lim, C.Z.H.; Yu, Q. Polycomb protein EZH2 regulates cancer cell fate decision in response to DNA damage. Cell Death Differ. 2011, 18, 1771–1779. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, M.E.; DuPrie, M.L.; Krueger, H.; Merajver, S.D.; Ventura, A.C.; Toy, K.A.; Kleer, C.G. Histone Methyltransferase EZH2 Induces Akt-Dependent Genomic Instability and BRCA1 Inhibition in Breast Cancer. Cancer Res. 2011, 71, 2360–2370. [Google Scholar] [CrossRef] [PubMed]
- Piunti, A.; Rossi, A.; Cerutti, A.; Albert, M.; Jammula, S.; Scelfo, A.; Cedrone, L.; Fragola, G.; Olsson, L.; Koseki, H.; Testa, G. Polycomb proteins control proliferation and transformation independently of cell cycle checkpoints by regulating DNA replication. Nat. Commun. 2014, 5, 3649. [Google Scholar] [CrossRef] [PubMed]
- Longo, V.D.; Fontana, L. Calorie restriction and cancer prevention: Metabolic and molecular mechanisms. Trends Pharmacol. Sci. 2010, 31, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Pietrocola, F.; Pol, J.; Vacchelli, E.; Rao, S.; Enot, D.P.; Baracco, E.E.; Levesque, S.; Castoldi, F.; Jacquelot, N.; Yamazaki, T.; et al. Caloric restriction mimetics enhance anticancer immunosurveillance. Cancer Cell 2016, 30, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Vermeij, W.P.; Dollé, M.E.T.; Reiling, E.; Jaarsma, D.; Payan-Gomez, C.; Bombardieri, C.R.; Wu, H.; Roks, A.J.; Botter, S.M.; Van Der Eerden, B.C.; Youssef, S.A. Restricted diet delays accelerated aging and genomic stress in DNA repair deficient mice. Nature 2016, 537, 427–431. [Google Scholar] [CrossRef] [PubMed]
- Beyaz, S.; Mana, M.D.; Roper, J.; Kedrin, D.; Saadatpour, A.; Hong, S.J.; Bauer-Rowe, K.E.; Xifaras, M.E.; Akkad, A.; Arias, E.; Pinello, L. High fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature 2016, 531, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Donohoe, C.L.; Lysaght, J.; O’Sullivan, J.; Reynolds, J.V. Emerging concepts linking obesity with the hallmarks of cancer. Trends Endocrinol. MeTable 2017, 28, 46–62. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Jin, Q.; Lee, J.E.; Su, I.H.; Ge, K. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 7317–7322. [Google Scholar] [CrossRef] [PubMed]
- Yi, S.A.; Um, S.H.; Lee, J.; Yoo, J.H.; Bang, S.Y.; Park, E.K.; Lee, M.G.; Nam, K.H.; Jeon, Y.J.; Park, J.W.; et al. S6K1 Phosphorylation of H2B Mediates EZH2 Trimethylation of H3: A Determinant of Early Adipogenesis. Mol. Cell 2016, 62, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Fontana, L.; Adelaiye, R.M.; Rastelli, A.L.; Miles, K.M.; Ciamporcero, E.; Longo, V.D.; Nguyen, H.; Vessella, R.; Pili, R. Dietary protein restriction inhibits tumor growth in human xenograft models. Oncotarget 2013, 4, 2451–2461. [Google Scholar] [CrossRef] [PubMed]
- Verrijzer, C.P. Sugarcoating Polycomb repression. Dev. Cell 2014, 31, 521–522. [Google Scholar] [CrossRef] [PubMed]
- Gambetta, M.C.; Muller, J. O-GlcNAcylation prevents aggregation of the Polycomb group repressor polyhomeotic. Dev. Cell 2014, 31, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Hanover, J.A.; Krause, M.W.; Love, D.C. Bittersweet memories: Linking metabolism to epigenetics through O-GlcNAcylation. Nat. Rev. Mol. Cell Biol. 2012, 13, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Dehennaut, V.; Leprince, D.; Lefebvre, T. O-GlcNAcylation, an Epigenetic Mark. Focus on the Histone Code, TET Family Proteins, and Polycomb Group Proteins. Front. Endocrinol. (Lausanne) 2014, 5, 155. [Google Scholar] [CrossRef] [PubMed]
- Dimri, M.; Bommi, P.V.; Sahasrabuddhe, A.A.; Khandekar, J.D.; Dimri, G.P. Dietary omega-3 polyunsaturated fatty acids suppress expression of EZH2 in breast cancer cells. Carcinogenesis 2010, 31, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cao, L.; Chen, J.; Song, S.; Lee, I.H.; Quijano, C.; Liu, H.; Keyvanfar, K.; Chen, H.; Cao, L.Y.; Ahn, B.H. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature 2009, 459, 387–392. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Bai, X.; Zhang, X.; Li, Z.; Tang, L.; Zhao, X.; Li, Z.; Ren, Y.; Wei, S.; Wang, Q.; Liu, C. Quantitative nuclear proteomics identifies that miR-137-mediated EZH2 reduction regulates resveratrol-induced apoptosis of neuroblastoma cells. Mol. Cell. Proteom. 2015, 14, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Ruan, T.; Liu, W.; Zhu, X.; Pan, J.; Lu, W.; Yan, C.; Tao, K.; Zhang, W.; Zhang, C. Effect and mechanism of curcumin on EZH2 - miR-101 regulatory feedback loop in multiple myeloma. Curr. Pharm. Des. 2017. [Google Scholar] [CrossRef]
- Wu, G.Q.; Chai, K.Q.; Zhu, X.M.; Jiang, H.; Wang, X.; Xue, Q.; Zheng, A.H.; Zhou, H.Y.; Chen, Y.; Chen, X.C.; Xiao, J.Y. Anti-cancer effects of curcumin on lung cancer through the inhibition of EZH2 and NOTCH1. Oncotarget 2016, 7, 26535–26550. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Huang, O.; Zhang, X.; Xie, Z.; Shen, A.; Liu, H.; Geng, M.; Shen, K. Curcumin induces cell death and restores tamoxifen sensitivity in the antiestrogen-resistant breast cancer cell lines MCF-7/LCC2 and MCF-7/LCC9. Molecules 2013, 18, 701–720. [Google Scholar] [CrossRef] [PubMed]
- Hua, W.F.; Fu, Y.S.; Liao, Y.J.; Xia, W.J.; Chen, Y.C.; Zeng, Y.X.; Kung, H.F.; Xie, D. Curcumin induces down-regulation of EZH2 expression through the MAPK pathway in MDA-MB-435 human breast cancer cells. Eur. J. Pharmacol. 2010, 637, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, S.R.; Balasubramanian, S.; Chew, Y.C.; Han, B.; Marquez, V.E.; Eckert, R.L. (−)-Epigallocatechin-3-gallate and DZNep reduce polycomb protein level via a proteasome-dependent mechanism in skin cancer cells. Carcinogenesis 2011, 32, 1525–1532. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, S.; Adhikary, G.; Eckert, R.L. The Bmi-1 polycomb protein antagonizes the (−)-epigallocatechin-3-gallate-dependent suppression of skin cancer cell survival. Carcinogenesis 2010, 31, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Shahabipour, F.; Caraglia, M.; Majeed, M.; Derosa, G.; Maffioli, P.; Sahebkar, A. Naturally occurring anti-cancer agents targeting EZH2. Cancer Lett. 2017, 400, 325–335. [Google Scholar] [CrossRef] [PubMed]
- McCabe, M.T.; Ott, H.M.; Ganji, G.; Korenchuk, S.; Thompson, C.; van Aller, G.S.; Liu, Y.; Graves, A.P.; Della Pietra, A., 3rd; Diaz, E.; et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature 2012, 492, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Yang, X.; Zhuang, L.; Jiang, X.; Chen, W.; Lee, P.L.; Karuturi, R.M.; Tan, P.B.O.; Liu, E.T.; Yu, Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes. Dev. 2007, 21, 1050–1063. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Bird, G.H.; Neff, T.; Guo, G.; Kerenyi, M.A.; Walensky, L.D.; Orkin, S.H. Targeted Disruption of the EZH2/EED Complex Inhibits EZH2-dependent Cancer. Nat. Chem. Biol. 2013, 9, 643–650. [Google Scholar] [CrossRef] [PubMed]
- Sashida, G.; Harada, H.; Matsui, H.; Oshima, M.; Yui, M.; Harada, Y.; Tanaka, S.; Mochizuki-Kashio, M.; Wang, C.; Saraya, A.; Muto, T. Ezh2 loss promotes development of myelodysplastic syndrome but attenuates its predisposition to leukaemic transformation. Nat. Commun. 2014, 5, 4177. [Google Scholar] [CrossRef] [PubMed]
- Comet, I.; Riising, E.M.; Leblanc, B.; Helin, K. Maintaining cell identity: PRC2-mediated regulation of transcription and cancer. Nat. Rev. Cancer 2016, 16, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Gibaja, V.; Shen, F.; Harari, J.; Korn, J.; Ruddy, D.; Saenz-Vash, V.; Zhai, H.; Rejtar, T.; Paris, C.G.; Yu, Z.; Lira, M. Development of secondary mutations in wild-type and mutant EZH2 alleles cooperates to confer resistance to EZH2 inhibitors. Oncogene 2016, 35, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Wassef, M.; Rodilla, V.; Teissandier, A.; Zeitouni, B.; Gruel, N.; Sadacca, B.; Irondelle, M.; Charruel, M.; Ducos, B.; Michaud, A.; Caron, M. Impaired PRC2 activity promotes transcriptional instability and favors breast tumorigenesis. Genes. Dev. 2015, 29, 2547–2562. [Google Scholar] [CrossRef] [PubMed]
| PRC2 Status | Associated Cancer Type | Experimental Therapeutic Approaches |
|---|---|---|
| EZH2 overexpression | Hematological malignancies [5] | miR-22 [6] |
| Pancreatic cancer [7,8] | DZNeP with gemcitabine [9] | |
| Chronic Pancreatitis [10] | ||
| Prostate cancer [11] | MicroRNA-101 [12] | |
| Breast cancer [13,14] | ||
| Bladder carcinoma [15,16,17] | NSC745885 [18], Gambogic acid and methyl jasmonate [19] | |
| Gastric cancer [20] | GSK126 [21] | |
| Lung cancer [22,23] | JQEZ5 [24], Schlafen11 [25] | |
| Hepatocellular carcinoma [26,27,28] | GSK343 [29,30] | |
| Glioblastoma multiforme [31] | DZNep, shRNA [32] | |
| Cervical Cancer [33,34] | ||
| Ovarian cancer [35] | ||
| Melanoma [36] | GSK126 [37], GSK503 [38] | |
| Soft Tissue Sarcoma [39,40] | ||
| Lymphoma, Mantle-Cell [41] | ||
| Colorectal cancer [42,43,44] | ||
| Retinoblastoma [45] | ||
| Tongue cancer [46,47] | ||
| EZH2 loss of function mutation | Peripheral nerve sheath tumors (MPNST) [48] | |
| Myeloproliferative neoplasms (MPNs) [49] | ||
| Myeloid –myelodysplastic malignancies [50,51] | ||
| Pediatric tumors of the central nervous system [52] | ||
| T cell acute lymphoblastic leukemia [53] | ||
| EZH2 gain of function mutation | Lymphomas EZH2-Tyr641 [54] EZH2-Ala677 [55] & Ala687 [56] | Small molecular inhibitor EI1 [57], GSK126 [58], EPZ005687 [58] |
| EED overexpession | Breast cancer [59] | |
| Colorectal cancer [60] | ||
| Hepatocellular carcinoma [61] | Wedelolactone [62] | |
| EED loss of function | Adenosquamous lung tumors [63] | |
| Malignant peripheral nerve sheath tumors [64] | ||
| SUZ12 overexpession | Breast cancer [61] | |
| Colorectal cancer [60,61] | ||
| Hepatocellular carcinoma [61] | ||
| Ovarian cancer [65] | ||
| Gastric Cancer [66,67] | ||
| Non-small cell lung cancer [68] | ||
| Bladder cancer [69] | ||
| SUZ12 loss of function | Malignant peripheral nerve sheath tumors [64] | |
| T cell acute lymphoblastic leukemia [53] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veneti, Z.; Gkouskou, K.K.; Eliopoulos, A.G. Polycomb Repressor Complex 2 in Genomic Instability and Cancer. Int. J. Mol. Sci. 2017, 18, 1657. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18081657
Veneti Z, Gkouskou KK, Eliopoulos AG. Polycomb Repressor Complex 2 in Genomic Instability and Cancer. International Journal of Molecular Sciences. 2017; 18(8):1657. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18081657
Chicago/Turabian StyleVeneti, Zoe, Kalliopi K. Gkouskou, and Aristides G. Eliopoulos. 2017. "Polycomb Repressor Complex 2 in Genomic Instability and Cancer" International Journal of Molecular Sciences 18, no. 8: 1657. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18081657