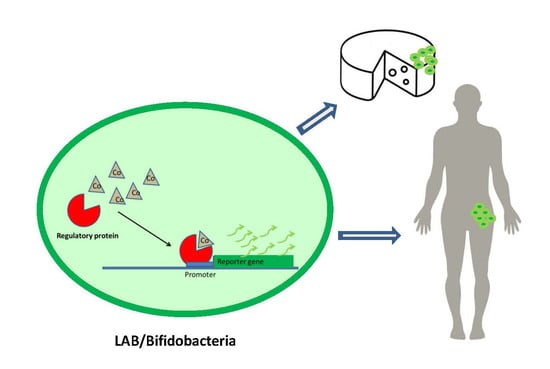
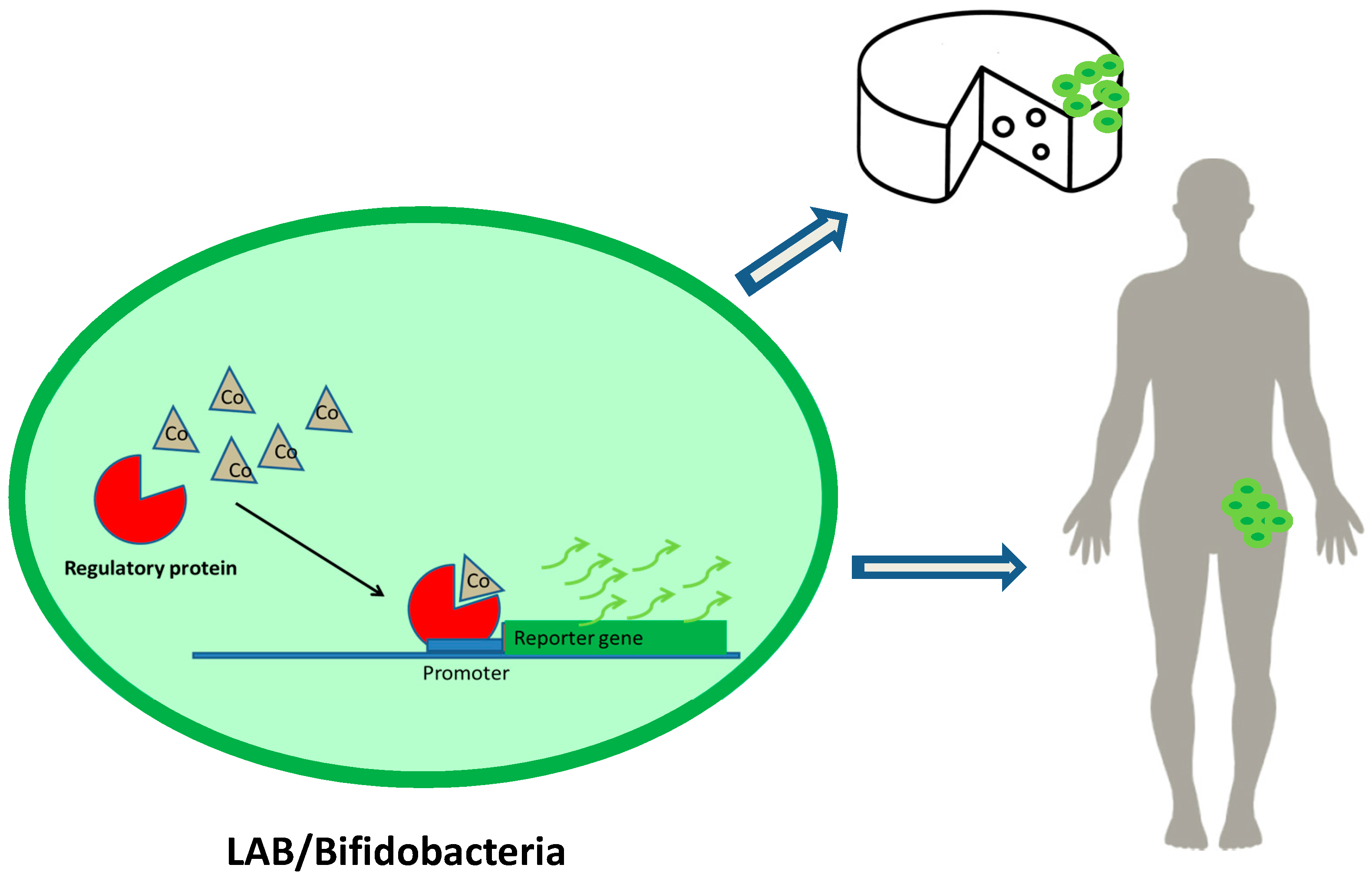
Fluorescent Lactic Acid Bacteria and Bifidobacteria as Vehicles of DNA Microbial Biosensors
Abstract
:1. Introduction
2. Reporter Genes Used in Lactic Acid Bacteria and Bifidobacteria
3. Identification of Biosensors
3.1. LAB and Bifidobacteria as Vehicles for Molecular Biosensors
3.2. Genetic Engineering for Molecular Biosensors
3.2.1. Food-Grade Cloning Vectors
3.2.2. Clustered Regularly Interspaced Short Palindromic Repeats
3.3. Microarray-RNAseq and Bidimensional Gel to Develop Molecular Biosensors
4. LAB and Bifidobacteria as Biosensors
5. Conclusions and Perspectives
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Park, M.; Tsai, S.-L.; Chen, W. Microbial biosensors: Engineered microorganisms as the sensing machinery. Sens. Basel 2013, 13, 5777–5795. [Google Scholar] [CrossRef] [PubMed]
- Sasson, V.; Shachrai, I.; Bren, A.; Dekel, E.; Alon, U. Mode of regulation and the insulation of bacterial gene expression. Mol. Cell 2012, 46, 399–407. [Google Scholar] [CrossRef] [PubMed]
- Galvão, T.C.; de Lorenzo, V. Transcriptional regulators à la carte: Engineering new effector specificities in bacterial regulatory proteins. Curr. Opin. Biotechnol. 2006, 17, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Landete, J.M. Effector molecules and regulatory proteins: Applications. Trends Biotechnol. 2016, 34, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Immonen, N.; Karp, M. Bioluminescence-based bioassays for rapid detection of nisin in food. Biosens. Bioelectron. 2007, 22, 1982–1987. [Google Scholar] [CrossRef] [PubMed]
- Laplace, J.M.; Hartke, A.; Giard, J.C.; Auffray, Y.J. Cloning, characterization and expression of an Enterococcus faecalis gene responsive to heavy metals. Appl. Microbiol. Biotechnol. 2000, 53, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Canbay, E.; Habip, A.; Kara, G.; Eren, Z.; Akyilmaz, E. A microbial biosensor based on Lactobacillus delbruecki sp. bacterial cells for simultaneous determination of lactic and pyruvic acid. Food Chem. 2015, 169, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Hutchens, M.; Luker, G.D. Applications of bioluminescence imaging to the study of infectious diseases. Cell Microbiol. 2007, 9, 2315–2322. [Google Scholar] [CrossRef] [PubMed]
- Justus, T.; Thomas, S.M. Evaluation of transcriptional fusions with green fluorescent protein versus luciferase as reporters in bacterial mutagenicity tests. Mutagenesis 1999, 14, 351–356. [Google Scholar] [CrossRef] [PubMed]
- Cronin, M.; Sleator, R.D.; Hill, C.; Fitzgerald, G.F.; van Sinderen, D. Development of a luciferase-based reporter system to monitor Bifidobacterium breve UCC2003 persistence in mice. BMC Microbiol. 2008, 8, 161. [Google Scholar] [CrossRef] [PubMed]
- Daniel, C.; Poiret, S.; Dennin, V.; Boutillier, D.; Pot, B. Bioluminescence imaging study of spatial and temporal persistence of Lactobacillus plantarum and Lactococcus lactis in living mice. Appl. Environ. Microbiol. 2013, 79, 1086–1094. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Campbell, R.E.; Ting, A.Y.; Tsien, R.Y. Creating new fluorescent probes for cell biology. Nat. Rev. Mol. Cell Biol. 2002, 3, 906–918. [Google Scholar] [CrossRef] [PubMed]
- Matz, M.V.; Fradkov, A.F.; Labas, Y.A.; Savitsky, A.P.; Zaraisky, A.G.; Markelov, M.L.; Lukyanov, S.A. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat. Biotechnol. 1999, 17, 969–973. [Google Scholar] [CrossRef] [PubMed]
- Shaner, N.C.; Campbell, R.E.; Steinbach, P.A.; Giepmans, B.N.; Palmer, A.E.; Tsien, R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004, 22, 1567–1572. [Google Scholar] [CrossRef] [PubMed]
- Shaner, N.C.; Steinbach, P.A.; Tsien, R.Y. A guide to choosing fluorescent proteins. Nat. Methods 2005, 2, 905–909. [Google Scholar] [CrossRef] [PubMed]
- Giepmans, B.N.G.; Adams, S.R.; Ellisman, M.H.; Tsien, R.Y. The fluorescent toolbox for assessing protein location and function. Science 2006, 312, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Giepmans, B.N.G. Bridging fluorescence microscopy and electron microscopy. Histochem. Cell Biol. 2008, 130, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Shcherbo, D.; Murphy, C.S.; Ermakova, G.V.; Solovieva, E.A.; Chepurnykh, T.V.; Shcheglov, A.S.; Verkhusha, V.V.; Pletnev, V.Z.; Hazelwood, K.L.; Roche, P.M.; et al. Far-red fluorescent tags for protein imaging in living tissues. Biochem. J. 2009, 418, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Tsien, R. The green fluorescent protein. Annu. Rev. Biochem. 1998, 67, 509–544. [Google Scholar] [CrossRef] [PubMed]
- Verkhusha, V.V.; Lukyanov, K.A. The molecular properties and applications of Anthozoa fluorescent proteins and chromoproteins. Nat. Biotechnol. 2004, 22, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Chudakov, D.M.; Lukyanov, S.; Lukyanov, K.A. Fluorescent proteins as a toolkit for in vivo imaging. Trends Biotechnol. 2005, 23, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Kremers, G.J.; Gilbert, S.G.; Cranfill, P.J.; Davidson, M.W.; Piston, D.W. Fluorescent proteins at a glance. J. Cell Sci. 2011, 124, 157–160. [Google Scholar] [CrossRef] [PubMed]
- Drepper, T.; Eggert, T.; Circolone, F.; Heck, A.; Krauss, U.; Guterl, J.K.; Wendorff, M.; Losi, A.; Gartner, W.; Jaeger, K.E. Reporter proteins for in vivo fluorescence without oxygen. Nat. Biotechnol. 2007, 25, 443–445. [Google Scholar] [CrossRef] [PubMed]
- Chapman, S.; Faulkner, C.; Kaiserli, E.; Garcia-Mata, C.; Savenkov, E.I.; Roberts, A.G.; Oparka, K.J.; Christie, J.M. The photoreversible fluorescent protein iLOV outperforms GFP as a reporter of plant virus infection. Proc. Natl. Acad. Sci. USA 2008, 105, 20038–20043. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Schroeder, C.M. Flavin-based fluorescent proteins: Emerging paradigms in biological imaging. Curr. Opin. Biotechnol. 2015, 31, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Landete, J.M.; Peirotén, A.; Rodríguez, E.; Margolles, A.; Medina, M.; Arqués, J.L. Anaerobic green fluorescent protein as a marker of Bifidobacterium strains. Int. J. Food Microbiol. 2014, 175, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Landete, J.M.; Langa, S.; Revilla, C.; Margolles, A.; Medina, M.; Arqués, J.L. Use of anaerobic green fluorescent protein versus green fluorescent protein as reporter in lactic acid bacteria. Appl. Microbiol. Biotechnol. 2015, 99, 6865–6877. [Google Scholar] [CrossRef] [PubMed]
- Geoffroy, M.C.; Guyard, C.; Quatannens, B.; Pavan, S.; Lange, M.; Mercenier, A. Use of green fluorescent protein to tag lactic acid bacterium strains under development as live vaccine vectors. Appl. Environ. Microbiol. 2000, 66, 383–391. [Google Scholar] [CrossRef] [PubMed]
- Bongaerts, R.J.M.; Hautefort, I.; Sidebotham, J.M.; Hinton, J.C.D. Green fluorescent protein as a marker for conditional gene expression in bacterial cells. Methods Enzymol. 2002, 358, 43–66. [Google Scholar] [PubMed]
- Wang, Y.P.; Wang, J.R.; Dai, W.L. Use of GFP to trace the colonization of Lactococcus lactis WH-C1 in the gastrointestinal tract of mice. J. Microbiol. Methods 2011, 86, 390–392. [Google Scholar] [CrossRef] [PubMed]
- García-Cayuela, T.; de Cadiñanos, L.P.; Mohedano, M.L.; de Palencia, P.F.; Boden, D.; Wells, J.; Peláez, C.; López, P.; Requena, T. Fluorescent protein vectors for promoter analysis in lactic acid bacteria and Escherichia coli. Appl. Microbiol. Biotechnol. 2012, 96, 171–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grimm, V.; Gleinser, M.; Neu, C.; Zhurina, D.; Riedel, C.U. Expression of fluorescent proteins in bifidobacteria for analysis of host-microbe interactions. Appl. Environ. Microbiol. 2014, 80, 2842–2850. [Google Scholar] [CrossRef] [PubMed]
- Tauer, C.; Heinl, S.; Egger, E.; Heiss, S.; Grabher, R. Tuning constitutive recombinant gene expression in Lactobacillus plantarum. Microb. Cell Fact. 2014, 13, 150. [Google Scholar] [CrossRef] [PubMed]
- Berlec, A.; Završnik, J.; Butinar, M.; Turk, B.; Štrukelj, B. In vivo imaging of Lactococcus lactis, Lactobacillus plantarum and Escherichia coli expressing infrared fluorescent protein in mice. Microb. Cell Fact. 2015, 14, 181. [Google Scholar] [CrossRef] [PubMed]
- Mohedano, M.L.; Garcia-Cayuela, T.; Perez-Ramos, A.; Gaiser, R.A.; Requena, T.; López, P. Construction and validation of a mCherry protein vector for promoter analysis in Lactobacillus acidophilus. J. Ind. Microbiol. Biotechnol. 2015, 42, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Montenegro-Rodríguez, C.; Peirotén, A.; Sanchez-Jimenez, A.; Arqués, J.L.; Landete, J.M. Analysis of gene expression of bifidobacteria using as the reporter an anaerobic fluorescent protein. Biotechnol. Lett. 2015, 37, 1405–1413. [Google Scholar] [CrossRef] [PubMed]
- Van Zyl, W.F.; Deane, S.M.; Dicks, L.M. Use of the mCherry fluorescent protein to study intestinal colonization by Enterococcus mundtii ST4SA and Lactobacillus plantarum 423 in mice. Appl. Environ. Microbiol. 2015, 81, 5993–6002. [Google Scholar] [CrossRef] [PubMed]
- Amaretti, A.; di Nunzio, M.; Pompei, A.; Raimondi, S.; Rossi, M.; Bordoni, A. Antioxidant properties of potentially probiotic bacteria in vitro and in vivo activities. Appl. Microbiol. Biotechnol. 2013, 97, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-H.; Gao, O.-Y.; Fang, J.Y. Meta-analysis of the efficacy and safety of Lactobacillus-containing and Bifidobacterium-containing probiotic compound preparation in Helicobacter pylori eradication therapy. J. Clin. Gastroenterol. 2013, 47, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Kassayová, M.; Bobrov, N.; Strojný, L.; Kisková, T.; Mikeš, J.; Demečková, V.; Orendáš, P.; Bojková, B.; Péč, M.; Kubatka, P.; et al. Preventive effects of probiotic bacteria Lactobacillus plantarum and dietary fiber in chemically-induced mammary carcinogenesis. Anticancer Res. 2014, 34, 4969–4975. [Google Scholar] [PubMed]
- Yang, S.; Li, W.; Challis, J.R.G.; Reid, G.; Kim, S.O.; Bocking, A.D. Probiotic Lactobacillus rhamnosus GR-1 supernatant prevents lipopolysaccharide-induced preterm birth and reduces inflammation in pregnant CD-1 mice. Am. J. Obst. Gynecol. 2014, 211, 44.e1–44.e12. [Google Scholar] [CrossRef] [PubMed]
- Oh, J.H.; van Pijkeren, J.P. CRISPR–Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res. 2014, 42, e131. [Google Scholar] [CrossRef] [PubMed]
- Landete, J.M. A review of food-grade cloning vectors in lactic acid bacteria: From the laboratory to their application. Crit. Rev. Biotechnol. 2016, 37, 296–308. [Google Scholar] [CrossRef] [PubMed]
- Holzapfel, W.H.; Haberer, P.; Geisen, R.; Björkroth, J.; Schillinger, U. Taxonomy and important features of probiotic microorganisms in food and nutrition. Am. J. Clin. Nutr. 2001, 73, 365s–373s. [Google Scholar] [PubMed]
- Charteris, W.P.; Kelly, P.M.; Morelli, P.M.; Collins, J.K. Selective detection, enumeration and identification of potentially probiotic Lactobacillus and Bifidobacterium species in mixed bacterial populations. Int. J. Food Microbiol. 1997, 35, 1–27. [Google Scholar] [CrossRef]
- Jankovic, I.; Sybesma, W.; Phothirath, P.; Ananta, E.; Mercenier, A. Application of probiotics in food products—Challenges and new approaches. Curr. Opin. Biotechnol. 2010, 21, 175–181. [Google Scholar] [CrossRef] [PubMed]
- Shareck, J.; Choi, Y.; Lee, B.; Miguez, C.B. Cloning vectors based on cryptic plasmids isolated from lactic acid bacteria: Their characteristics and potential applications in biotechnology. Crit. Rev. Biotechnol. 2004, 24, 155–208. [Google Scholar] [CrossRef] [PubMed]
- De Vos, W.M. Safe and sustainable systems for food-grade fermentations by genetically modified lactic acid bacteria. Int. Dairy J. 1999, 9, 3–10. [Google Scholar] [CrossRef]
- Lu, W.; Kong, J.; Kong, W. Construction and application of a foodgrade expression system for Lactococcus lactis. Mol. Biotechnol. 2013, 54, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Takala, T.M.; Qiao, M.; Xu, H.; Saris, P.E.J. Nisin-selectable food-grade secretion vector for Lactococcus lactis. Biotechnol. Lett. 2011, 33, 797–803. [Google Scholar] [CrossRef] [PubMed]
- Allison, G.E.; Klaenhammer, T.R. Functional analysis of the gene encoding immunity to lactacin F, lafI, and its use as a Lactobacillus-specific, food-grade genetic marker. Appl. Environ. Microbiol. 1996, 62, 4450–4460. [Google Scholar] [PubMed]
- Sorek, R.; Kunin, V.; Hugenholtz, P. CRISPR—A widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 2008, 6, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Hovath, P.; Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Sander, J.D.; Joung, J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014, 32, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Stefanovic, E.; Fitzgerald, G.; McAuliffe, O. Advances in the genomics and metabolomics of dairy lactobacilli: A review. Food Microbiol. 2017, 61, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Van Pijkeren, J.P.; Neoh, K.M.; Sirias, D.; Findley, A.S.; Britton, R.A. Exploring optimization parameters to increase ssDNA recombineering in Lactococcus lactis and Lactobacillus reuteri. Bioengineered 2012, 3, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Van Pijkeren, J.P.; Britton, R.A. High efficiency recombineering in lactic acid bacteria. Nucleic Acids Res. 2012, 40, e76. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Bikard, D.; Cox, D.; Zhang, F.; Marraffini, L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013, 31, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Portmann, R.; Magnani, D.; Stoyanov, J.V.; Schmechel, A.; Multhaup, G.; Solioz, M. Interaction kinetics of the copper-responsive CopY repressor with the cop promoter of Enterococcus hirae. J. Biol. Inorg. Chem. 2004, 9, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Rogers, J.K.; Guzman, C.D.; Taylor, N.D.; Raman, S.; Anderson, K.; Church, G.M. Synthetic biosensors for precise gene control and real-time monitoring of metabolites. Nucleic Acids Res. 2015, 43, 7648–7660. [Google Scholar] [CrossRef] [PubMed]
- Koskenniemi, K.; Koponen, J.; Kankainen, M.; Savijoki, K.; Tynkkynen, S.; de Vos, W.M.; Kalkkinen, N.; Varmanen, P. Proteome analysis of Lactobacillus rhamnosus GG using 2-D DIGE and Mass Spectrometry shows differential protein production in laboratory and industrial-type growth media. Proteome Res. 2009, 8, 4993–5007. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Zhang, W.; Sun, T.; Wu, J.; Yue, X.; Meng, H.; Zhang, H. Proteomic analysis of responses of a new probiotic bacterium Lactobacillus casei Zhang to low acid stress. Int. J. Food Microbiol. 2011, 147, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Magnani, D.; Barré, O.; Gerber, S.D.; Soliozm, M. Characterization of the CopR Regulon of Lactococcus lactis IL1403. J. Bacteriol. 2008, 190, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Virolainen, N.; Guglielmetti, S.; Arioli, S.; Karp, M. Bioluminescence-based identification of nisin producers—A rapid and simple screening method for nisinogenic bacteria in food samples. Int. J. Food Microbiol. 2012, 158, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Hilmi, H.T.A.; Hakkila, K.; Saris, P.E.J. Isolation of sensitive nisin-sensing GFPuv bioassay Lactococcus lactis strains using FACS. Biotechnol. Lett. 2009, 31, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Oozeer, R.; Furet, J.P.; Goupil-Feuillerat, N.; Anba, J.; Mengaud, J.; Corthier, G. Differential activities of four Lactobacillus casei promoters during bacterial transit through the gastrointestinal tracts of human-microbiota-associated mice. Appl. Environ. Microbiol. 2005, 71, 1356–1363. [Google Scholar] [CrossRef] [PubMed]
- Ravnikar, M.; Štrukelj, B.; Obermajer, N.; Lunder, M.; Berlec, A. Engineered lactic acid bacterium Lactococcus lactis capable of binding antibodies and Tumor Necrosis Factor Alpha. Appl. Environ. Microbiol. 2010, 76, 6928–6932. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.; Dosoky, N.S.; Berdiev, B.K.; Williams, J.D. Electrochemical impedance spectroscopy for black lipid membranes fused with channel protein supported on solid-state nanopore. Eur. Biophys. J. 2016, 45, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.; Misra, S.K.; Schwartz-Duval, A.S.; Daza, E.; Ostadhossein, F.; Bowman, M.; Jain, A.; Taylor, G.; McDonagh, D.; Labriola, L.T.; et al. Real-time monitoring of post-surgical and post-traumatic eye injuries using multilayered electrical biosensor chip. ACS Appl. Mater. Interfaces 2017, 9, 8609–8622. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.; Misra, S.K.; Wang, Z.; Daza, E.; Schwartz-Duval, A.S.; Kus, J.M.; Pan, D.; Pan, D. Paper-based analytical biosensor chip designed from graphene-nanoplatelet-amphiphilic-diblock-co-polymer composite for cortisol detection in human saliva. Anal. Chem. 2017, 89, 2107–2115. [Google Scholar] [CrossRef] [PubMed]
- Linares, D.M.; Perez, M.; del Rio, B.; Redruello, B.; Martin, M.C.; Fernández, M.; Alvarez, M.A. An agmatine-inducible system for the expression of recombinant proteins in Enterococcus faecalis. Microb. Cell Fact. 2014, 13, 169. [Google Scholar] [CrossRef] [PubMed]
- Guglielmetti, S.; Ciranna, A.; Mora, D.; Parini, C.; Karp, M. Construction, characterization and exemplificative application of bioluminescent Bifidobacterium longum biovar longum. Int. J. Food Microbiol. 2008, 124, 285–290. [Google Scholar] [CrossRef] [PubMed]


| Bacteria | Reporters | Promoters | Effector | Reference |
|---|---|---|---|---|
| Lactococcus lactis NZ9800/NZ9000 | lux reporter system | Pnis | Nisin in food samples | [5] |
| Enterococcus faecalis JH2-2 | 32P-labeled probe of csrA cDNA | csrA | Heavy metals | [6] |
| Enterococcus hirae | lux reporter system | cop | Copper | [59] |
| Lactococcus lactis IL1403 | - | YaiA, YtjD, LctO | Copper | [63] |
| Lactococcus lactis NZ9800 | lux reporter system | Pnis | Nisin producers | [64] |
| Lactococcus lactis LAC275 | GFP | Pnis | Nisin | [65] |
| Lactobacillus casei DN-114 001 | lux reporter system | ccpA, dlt, ldh, lacT | Changes in the gastrointestinal tract | [66] |
| Enterococcus faecalis | GFP | aguB | Agmantine | [68] |
| Bifidobacterium longum | lux reporter system | phage T5 promoter | Carbohydrates | [69] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Landete, J.M.; Arqués, J.L. Fluorescent Lactic Acid Bacteria and Bifidobacteria as Vehicles of DNA Microbial Biosensors. Int. J. Mol. Sci. 2017, 18, 1728. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18081728
Landete JM, Arqués JL. Fluorescent Lactic Acid Bacteria and Bifidobacteria as Vehicles of DNA Microbial Biosensors. International Journal of Molecular Sciences. 2017; 18(8):1728. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18081728
Chicago/Turabian StyleLandete, José María, and Juan Luis Arqués. 2017. "Fluorescent Lactic Acid Bacteria and Bifidobacteria as Vehicles of DNA Microbial Biosensors" International Journal of Molecular Sciences 18, no. 8: 1728. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18081728