To Wnt or Lose: The Missing Non-Coding Linc in Colorectal Cancer
Abstract
:1. Introduction
2. Wnt: The Initiating and Driving Force of Colorectal Cancer (CRC)
3. LncRNAs: The Emerging Dark Matters That Matter
4. LncRNAs in Wnt Signaling and CRC
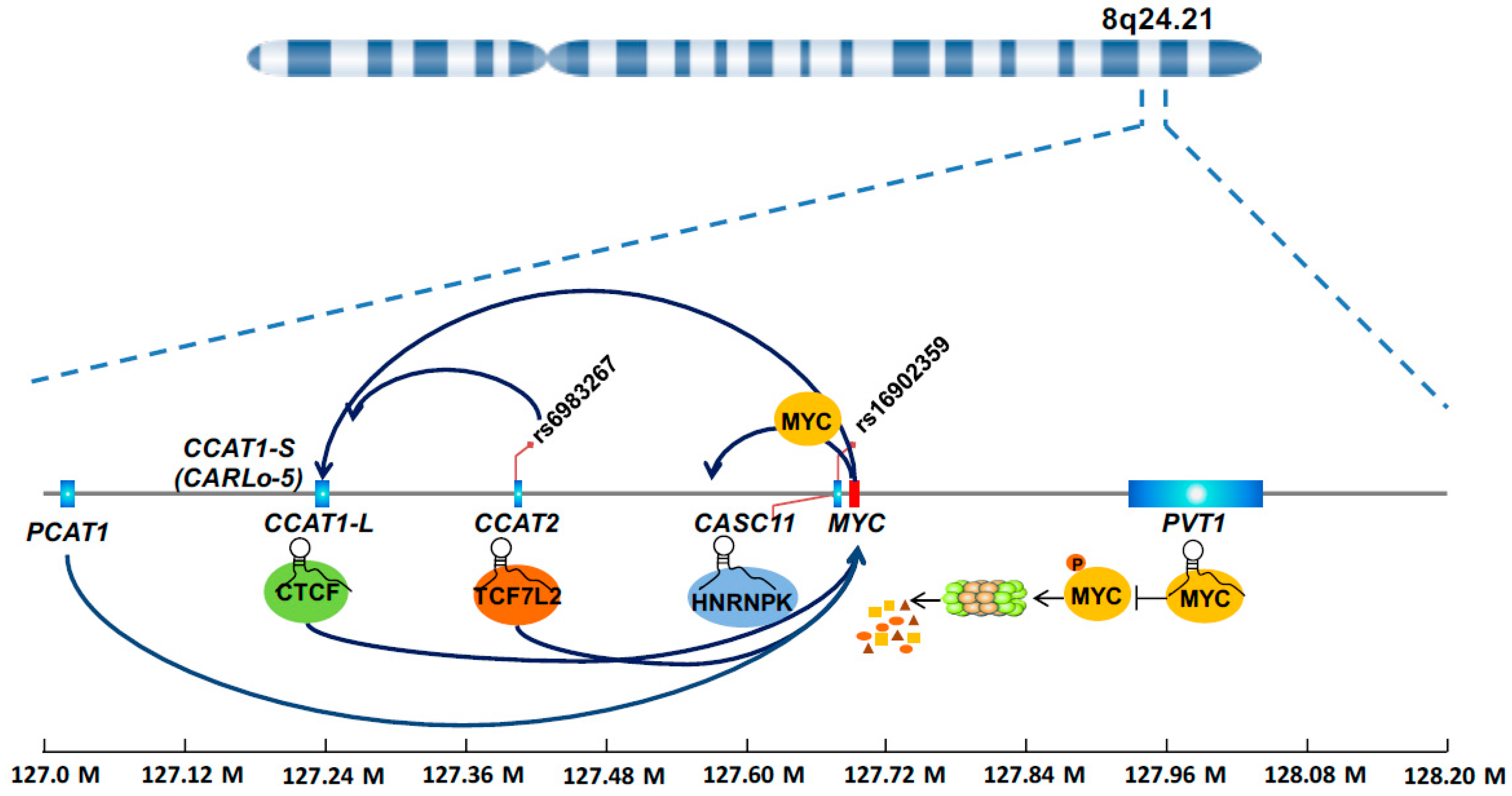
4.1. LncRNAs from 8q24 Region
4.1.1. CCAT1
4.1.2. CCAT2
4.1.3. CASC11
4.1.4. PVT1
4.1.5. PCAT1
4.2. CRC Stem Cell—Associated LncRNAs
4.2.1. Lnc34a
4.2.2. RBM5-AS1
4.2.3. WiNTRLINC1
4.3. Others
4.3.1. H19
4.3.2. CCAL
4.3.3. CTD903
4.3.4. ASBEL
4.3.5. MYU
5. Potential Clinical Application
6. Conclusions
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fedewa, S.A.; Ahnen, D.J.; Meester, R.G. S.; Barzi, A.; Jemal, A. Colorectal cancer statistics, 2017. CA Cancer J. Clin. 2017, 67, 177–193. [Google Scholar] [CrossRef] [PubMed]
- Goss, K.H.; Groden, J. Biology of the adenomatous polyposis coli tumor suppressor. J. Clin. Oncol. 2000, 18, 1967–1979. [Google Scholar] [CrossRef] [PubMed]
- Giles, R.H.; van Es, J.H.; Clevers, H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim. Biophys. Acta 2003, 1653, 1–24. [Google Scholar] [CrossRef]
- Groden, J.; Thliveris, A.; Samowitz, W.; Carlson, M.; Gelbert, L.; Albertsen, H.; Joslyn, G.; Stevens, J.; Spirio, L.; Robertson, M.; et al. Identification and characterization of the familial adenomatous polyposis coli gene. Cell 1991, 66, 589–600. [Google Scholar] [CrossRef]
- Nagel, R.; le Sage, C.; Diosdado, B.; van der Waal, M.; Oude Vrielink, J.A.; Bolijn, A.; Meijer, G.A.; Agami, R. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008, 68, 5795–5802. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.H.; Kim, H.S.; Kim, N.G.; Lee, I.; Choi, H.S.; Li, X.Y.; Kang, S.E.; Cha, S.Y.; Ryu, J.K.; Na, J.M.; et al. p53 and microRNA-34 are suppressors of canonical Wnt signaling. Sci. Signal. 2011, 4, ra71. [Google Scholar] [CrossRef] [PubMed]
- Hwang, W.L.; Jiang, J.K.; Yang, S.H.; Huang, T.S.; Lan, H.Y.; Teng, H.W.; Yang, C.Y.; Tsai, Y.P.; Lin, C.H.; Wang, H.W.; et al. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat. Cell Biol. 2014, 16, 268–280. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Smolle, M.; Uranitsch, S.; Gerger, A.; Pichler, M.; Haybaeck, J. Current status of long non-coding RNAs in human cancer with specific focus on colorectal cancer. Int. J. Mol. Sci. 2014, 15, 13993–14013. [Google Scholar] [CrossRef] [PubMed]
- Nusse, R.; Clevers, H. Wnt/β-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017, 169, 985–999. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Non-coding RNAs in human disease. Nat. Rev. Genet. 2011, 12, 861–874. [Google Scholar] [CrossRef] [PubMed]
- Nusse, R.; Varmus, H.E. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell 1982, 31, 99–109. [Google Scholar] [CrossRef]
- Rijsewijk, F.; Schuermann, M.; Wagenaar, E.; Parren, P.; Weigel, D.; Nusse, R. The Drosophila homolog of the mouse mammary oncogene int-1 is identical to the segment polarity gene wingless. Cell 1987, 50, 649–657. [Google Scholar] [CrossRef]
- Kikuchi, A.; Yamamoto, H.; Sato, A.; Matsumoto, S. New insights into the mechanism of Wnt signaling pathway activation. Int. Rev. Cell Mol. Biol. 2011, 291, 21–71. [Google Scholar] [PubMed]
- Hart, M.J.; de los Santos, R.; Albert, I.N.; Rubinfeld, B.; Polakis, P. Downregulation of β-catenin by human Axin and its association with the APC tumor suppressor, β-catenin and GSK3β. Curr. Biol. 1998, 8, 573–581. [Google Scholar] [CrossRef]
- Kishida, S.; Yamamoto, H.; Ikeda, S.; Kishida, M.; Sakamoto, I.; Koyama, S.; Kikuchi, A. Axin, a negative regulator of the Wnt signaling pathway, directly interacts with adenomatous polyposis coli and regulates the stabilization of β-catenin. J. Biol. Chem. 1998, 273, 10823–10826. [Google Scholar] [CrossRef] [PubMed]
- Brannon, M.; Gomperts, M.; Sumoy, L.; Moon, R.T.; Kimelman, D. A β-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev. 1997, 11, 2359–2370. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar]
- Powell, S.M.; Zilz, N.; Beazer-Barclay, Y.; Bryan, T.M.; Hamilton, S.R.; Thibodeau, S.N.; Vogelstein, B.; Kinzler, K.W. APC mutations occur early during colorectal tumorigenesis. Nature 1992, 359, 235–237. [Google Scholar] [CrossRef] [PubMed]
- Burt, R.W.; Leppert, M.F.; Slattery, M.L.; Samowitz, W.S.; Spirio, L.N.; Kerber, R.A.; Kuwada, S.K.; Neklason, D.W.; Disario, J.A.; Lyon, E.; et al. Genetic testing and phenotype in a large kindred with attenuated familial adenomatous polyposis. Gastroenterology 2004, 127, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Morin, P.J.; Sparks, A.B.; Korinek, V.; Barker, N.; Clevers, H.; Vogelstein, B.; Kinzler, K.W. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science 1997, 275, 1787–1790. [Google Scholar] [CrossRef] [PubMed]
- Korinek, V.; Barker, N.; Morin, P.J.; van Wichen, D.; de Weger, R.; Kinzler, K.W.; Vogelstein, B.; Clevers, H. Constitutive transcriptional activation by a β-catenin-Tcf complex in APC−/− colon carcinoma. Science 1997, 275, 1784–1787. [Google Scholar] [CrossRef] [PubMed]
- Vermeulen, L.; de Sousa, E.M.F.; van der Heijden, M.; Cameron, K.; de Jong, J.H.; Borovski, T.; Tuynman, J.B.; Todaro, M.; Merz, C.; Rodermond, H.; et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat. Cell Biol. 2010, 12, 468–476. [Google Scholar] [CrossRef] [PubMed]
- Dow, L.E.; O’Rourke, K.P.; Simon, J.; Tschaharganeh, D.F.; van Es, J.H.; Clevers, H.; Lowe, S.W. Apc Restoration Promotes Cellular Differentiation and Reestablishes Crypt Homeostasis in Colorectal Cancer. Cell 2015, 161, 1539–1552. [Google Scholar] [CrossRef] [PubMed]
- Tetsu, O.; McCormick, F. β-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 1999, 398, 422–426. [Google Scholar] [PubMed]
- He, T.C.; Sparks, A.B.; Rago, C.; Hermeking, H.; Zawel, L.; da Costa, L.T.; Morin, P.J.; Vogelstein, B.; Kinzler, K.W. Identification of c-MYC as a target of the APC pathway. Science 1998, 281, 1509–1512. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Vincent, K.; Pichler, M.; Fodde, R.; Berindan-Neagoe, I.; Slack, F.J.; Calin, G.A. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene 2015, 34, 5003–5011. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Beroukhim, R.; Mermel, C.H.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M.; et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010, 463, 899–905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, L.; Qi, H.; Luo, F.; Xu, H.; Ling, M.; Qin, Y.; Yang, P.; Liu, X.; Yang, Q.; Xue, J.; et al. Feedback circuitry via let-7c between lncRNA CCAT1 and c-Myc is involved in cigarette smoke extract-induced malignant transformation of HBE cells. Oncotarget 2017, 8, 19285–19297. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhou, C.; Chang, Y.; Zhang, Z.; Hu, Y.; Zhang, F.; Lu, Y.; Zheng, L.; Zhang, W.; Li, X.; Li, X. Long non-coding RNA CASC11 interacts with hnRNP-K and activates the WNT/β-catenin pathway to promote growth and metastasis in colorectal cancer. Cancer Lett. 2016, 376, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Tseng, Y.Y.; Moriarity, B.S.; Gong, W.; Akiyama, R.; Tiwari, A.; Kawakami, H.; Ronning, P.; Reuland, B.; Guenther, K.; Beadnell, T.C.; et al. PVT1 dependence in cancer with MYC copy-number increase. Nature 2014, 512, 82–86. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Cui, R.; Jeon, Y.J.; Lee, J.H.; Lee, J.H.; Sim, H.; Park, J.K.; Fadda, P.; Tili, E.; Nakanishi, H.; et al. Long-range interaction and correlation between MYC enhancer and oncogenic long noncoding RNA CARLo-5. Proc. Natl. Acad. Sci. USA 2014, 111, 4173–4178. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Spizzo, R.; Atlasi, Y.; Nicoloso, M.; Shimizu, M.; Redis, R.S.; Nishida, N.; Gafa, R.; Song, J.; Guo, Z.; et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013, 23, 1446–1461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, N.; Penn, L.Z. Reflecting on 25 years with MYC. Nat. Rev. Cancer 2008, 8, 976–990. [Google Scholar] [CrossRef] [PubMed]
- Tenesa, A.; Farrington, S.M.; Prendergast, J.G.; Porteous, M.E.; Walker, M.; Haq, N.; Barnetson, R.A.; Theodoratou, E.; Cetnarskyj, R.; Cartwright, N.; et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on 11q23 and replicates risk loci at 8q24 and 18q21. Nat. Genet. 2008, 40, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, I.; Webb, E.; Carvajal-Carmona, L.; Broderick, P.; Kemp, Z.; Spain, S.; Penegar, S.; Chandler, I.; Gorman, M.; Wood, W.; et al. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21. Nat. Genet. 2007, 39, 984–988. [Google Scholar] [CrossRef] [PubMed]
- Zanke, B.W.; Greenwood, C.M.; Rangrej, J.; Kustra, R.; Tenesa, A.; Farrington, S.M.; Prendergast, J.; Olschwang, S.; Chiang, T.; Crowdy, E.; et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat. Genet. 2007, 39, 989–994. [Google Scholar] [CrossRef] [PubMed]
- Pomerantz, M.M.; Ahmadiyeh, N.; Jia, L.; Herman, P.; Verzi, M.P.; Doddapaneni, H.; Beckwith, C.A.; Chan, J.A.; Hills, A.; Davis, M.; et al. The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat. Genet. 2009, 41, 882–884. [Google Scholar] [CrossRef] [PubMed]
- Tuupanen, S.; Turunen, M.; Lehtonen, R.; Hallikas, O.; Vanharanta, S.; Kivioja, T.; Bjorklund, M.; Wei, G.; Yan, J.; Niittymaki, I.; et al. The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat. Genet. 2009, 41, 885–890. [Google Scholar] [CrossRef] [PubMed]
- Hnisz, D.; Abraham, B.J.; Lee, T.I.; Lau, A.; Saint-Andre, V.; Sigova, A.A.; Hoke, H.A.; Young, R.A. Super-enhancers in the control of cell identity and disease. Cell 2013, 155, 934–947. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.; Wu, F.; Gao, F.; Qing, X.; Shao, Z. Prognostic value of long non-coding RNA CCAT1 expression in patients with cancer: A meta-analysis. PLoS ONE 2017, 12, e0179346. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.F.; Yin, Q.F.; Chen, T.; Zhang, Y.; Zhang, X.O.; Wu, Z.; Zhang, S.; Wang, H.B.; Ge, J.; Lu, X.; et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014, 24, 513–531. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Liu, C.G.; Ferracin, M.; Hyslop, T.; Spizzo, R.; Sevignani, C.; Fabbri, M.; Cimmino, A.; Lee, E.J.; Wojcik, S.E.; et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 2007, 12, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Haiman, C.A.; Le Marchand, L.; Yamamato, J.; Stram, D.O.; Sheng, X.; Kolonel, L.N.; Wu, A.H.; Reich, D.; Henderson, B.E. A common genetic risk factor for colorectal and prostate cancer. Nat. Genet. 2007, 39, 954–956. [Google Scholar] [CrossRef] [PubMed]
- Yeager, M.; Orr, N.; Hayes, R.B.; Jacobs, K.B.; Kraft, P.; Wacholder, S.; Minichiello, M.J.; Fearnhead, P.; Yu, K.; Chatterjee, N.; et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat. Genet. 2007, 39, 645–649. [Google Scholar] [CrossRef] [PubMed]
- Sur, I.K.; Hallikas, O.; Vaharautio, A.; Yan, J.; Turunen, M.; Enge, M.; Taipale, M.; Karhu, A.; Aaltonen, L.A.; Taipale, J. Mice lacking a Myc enhancer that includes human SNP rs6983267 are resistant to intestinal tumors. Science 2012, 338, 1360–1363. [Google Scholar] [CrossRef] [PubMed]
- Redis, R.S.; Vela, L.E.; Lu, W.; Ferreira de Oliveira, J.; Ivan, C.; Rodriguez-Aguayo, C.; Adamoski, D.; Pasculli, B.; Taguchi, A.; Chen, Y.; et al. Allele-Specific Reprogramming of Cancer Metabolism by the Long Non-coding RNA CCAT2. Mol. Cell 2016, 61, 520–534. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.; Liang, H.; Cui, X.; Han, C.; Hao, C.; Huo, K. Long noncoding RNA CCAT2 can predict metastasis and a poor prognosis: A meta-analysis. Clin. Chim. Acta 2017, 468, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, T.; Matsuyama, T.; Toiyama, Y.; Takahashi, N.; Ishikawa, T.; Uetake, H.; Yamada, Y.; Kusunoki, M.; Calin, G.; Goel, A. CCAT1 and CCAT2 long noncoding RNAs, located within the 8q.24.21 ’gene desert’, serve as important prognostic biomarkers in colorectal cancer. Ann. Oncol. 2017, 28, 1882–1888. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.H.; Fang, H.; Ji, C.X.; Xie, H.; Xiao, B.; Zhu, X.G. Long noncoding RNA CCAT2 can predict metastasis and poor prognosis: A meta-analysis. Clin. Chim. Acta 2017, 466, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.; Hou, Y.C.; Fu, L.N.; Wang, Y.Q.; Liu, Q.Q.; Xiong, H.; Chen, Y.X.; Fang, J.Y. Long Noncoding RNA CCAT2 as a Potential Novel Biomarker to Predict the Clinical Outcome of Cancer Patients: A Meta-Analysis. J. Cancer 2017, 8, 1498–1506. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Sawada, G.; Kurashige, J.; Uchi, R.; Matsumura, T.; Ueo, H.; Takano, Y.; Eguchi, H.; Sudo, T.; Sugimachi, K.; et al. Amplification of PVT-1 is involved in poor prognosis via apoptosis inhibition in colorectal cancers. Br. J. Cancer 2014, 110, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Huppi, K.; Volfovsky, N.; Runfola, T.; Jones, T.L.; Mackiewicz, M.; Martin, S.E.; Mushinski, J.F.; Stephens, R.; Caplen, N.J. The identification of microRNAs in a genomically unstable region of human chromosome 8q24. Mol. Cancer Res. 2008, 6, 212–221. [Google Scholar] [CrossRef] [PubMed]
- Prensner, J.R.; Iyer, M.K.; Balbin, O.A.; Dhanasekaran, S.M.; Cao, Q.; Brenner, J.C.; Laxman, B.; Asangani, I.A.; Grasso, C.S.; Kominsky, H.D.; et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat. Biotechnol. 2011, 29, 742–749. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Ahmed, M.; Zhang, F.; Yao, C.Q.; Li, S.; Liang, Y.; Hua, J.; Soares, F.; Sun, Y.; Langstein, J.; et al. Modulation of long noncoding RNAs by risk SNPs underlying genetic predispositions to prostate cancer. Nat. Genet. 2016, 48, 1142–1150. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Chen, Y.; Liao, X.; Liu, D.; Li, F.; Ruan, H.; Jia, W. Overexpression of long noncoding RNA PCAT-1 is a novel biomarker of poor prognosis in patients with colorectal cancer. Med. Oncol. 2013, 30, 588. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.; Liu, X.; Tang, Y.; Zhao, Z.; Zhang, J.; Feng, Y. Down regulation of the long non-coding RNA PCAT-1 induced growth arrest and apoptosis of colorectal cancer cells. Life Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Ordonez-Moran, P.; Dafflon, C.; Imajo, M.; Nishida, E.; Huelsken, J. HOXA5 Counteracts Stem Cell Traits by Inhibiting Wnt Signaling in Colorectal Cancer. Cancer Cell 2015, 28, 815–829. [Google Scholar] [CrossRef] [PubMed]
- Barker, N.; van Es, J.H.; Kuipers, J.; Kujala, P.; van den Born, M.; Cozijnsen, M.; Haegebarth, A.; Korving, J.; Begthel, H.; Peters, P.J.; Clevers, H. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 2007, 449, 1003–1007. [Google Scholar] [CrossRef] [PubMed]
- Van der Flier, L.G.; van Gijn, M.E.; Hatzis, P.; Kujala, P.; Haegebarth, A.; Stange, D.E.; Begthel, H.; van den Born, M.; Guryev, V.; Oving, I.; et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell 2009, 136, 903–912. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Yang, Y.; Wang, F.; Moyer, M.P.; Wei, Q.; Zhang, P.; Yang, Z.; Liu, W.; Zhang, H.; Chen, N.; et al. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/β-catenin signaling pathway via suppression of activator protein 2α. Gut 2016, 65, 1494–1504. [Google Scholar] [CrossRef] [PubMed]
- Kendziorra, E.; Ahlborn, K.; Spitzner, M.; Rave-Frank, M.; Emons, G.; Gaedcke, J.; Kramer, F.; Wolff, H.A.; Becker, H.; Beissbarth, T.; et al. Silencing of the Wnt transcription factor TCF4 sensitizes colorectal cancer cells to (chemo-) radiotherapy. Carcinogenesis 2011, 32, 1824–1831. [Google Scholar] [CrossRef] [PubMed]
- Li, X.J.; Ren, Z.J.; Tang, J.H. MicroRNA-34a: A potential therapeutic target in human cancer. Cell Death Dis. 2014, 5, e1327. [Google Scholar] [CrossRef] [PubMed]
- Hermeking, H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010, 17, 193–199. [Google Scholar] [CrossRef] [PubMed]
- Hashimi, S.T.; Fulcher, J.A.; Chang, M.H.; Gov, L.; Wang, S.; Lee, B. MicroRNA profiling identifies miR-34a and miR-21 and their target genes JAG1 and WNT1 in the coordinate regulation of dendritic cell differentiation. Blood 2009, 114, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Fu, H.; Liu, Q.; Tie, Y.; Zhu, J.; Xing, R.; Sun, Z.; Zheng, X. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008, 582, 1564–1568. [Google Scholar] [CrossRef] [PubMed]
- Bu, P.; Wang, L.; Chen, K.Y.; Srinivasan, T.; Murthy, P.K.; Tung, K.L.; Varanko, A.K.; Chen, H.J.; Ai, Y.; King, S.; et al. A miR-34a-Numb Feedforward Loop Triggered by Inflammation Regulates Asymmetric Stem Cell Division in Intestine and Colon Cancer. Cell Stem Cell 2016, 18, 189–202. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Bu, P.; Ai, Y.; Srinivasan, T.; Chen, H.J.; Xiang, K.; Lipkin, S.M.; Shen, X. A long non-coding RNA targets microRNA miR-34a to regulate colon cancer stem cell asymmetric division. Elife 2016, 5, e14620. [Google Scholar] [CrossRef] [PubMed]
- Di Cecilia, S.; Zhang, F.; Sancho, A.; Li, S.; Aguilo, F.; Sun, Y.; Rengasamy, M.; Zhang, W.; del Vecchio, L.; Salvatore, F.; et al. RBM5-AS1 Is Critical for Self-Renewal of Colon Cancer Stem-like Cells. Cancer Res. 2016, 76, 5615–5627. [Google Scholar] [CrossRef] [PubMed]
- Giakountis, A.; Moulos, P.; Zarkou, V.; Oikonomou, C.; Harokopos, V.; Hatzigeorgiou, A.G.; Reczko, M.; Hatzis, P. A Positive Regulatory Loop between a Wnt-Regulated Non-coding RNA and ASCL2 Controls Intestinal Stem Cell Fate. Cell Rep. 2016, 15, 2588–2596. [Google Scholar] [CrossRef] [PubMed]
- Brannan, C.I.; Dees, E.C.; Ingram, R.S.; Tilghman, S.M. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 1990, 10, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shields, T.; Crenshaw, T.; Hao, Y.; Moulton, T.; Tycko, B. Imprinting of human H19: Allele-specific CpG methylation, loss of the active allele in Wilms tumor, and potential for somatic allele switching. Am. J. Hum. Genet. 1993, 53, 113–124. [Google Scholar] [PubMed]
- Zhang, Y.; Tycko, B. Monoallelic expression of the human H19 gene. Nat. Genet. 1992, 1, 40–44. [Google Scholar] [CrossRef] [PubMed]
- Raveh, E.; Matouk, I.J.; Gilon, M.; Hochberg, A. The H19 Long non-coding RNA in cancer initiation, progression and metastasis—A proposed unifying theory. Mol. Cancer 2015, 14, 184. [Google Scholar] [CrossRef] [PubMed]
- Medrzycki, M.; Zhang, Y.; Zhang, W.; Cao, K.; Pan, C.; Lailler, N.; McDonald, J.F.; Bouhassira, E.E.; Fan, Y. Histone H1.3 suppresses H19 noncoding RNA expression and cell growth of ovarian cancer cells. Cancer Res. 2014, 74, 6463–6473. [Google Scholar] [CrossRef] [PubMed]
- Moulton, T.; Crenshaw, T.; Hao, Y.; Moosikasuwan, J.; Lin, N.; Dembitzer, F.; Hensle, T.; Weiss, L.; McMorrow, L.; Loew, T.; et al. Epigenetic lesions at the H19 locus in Wilms’ tumour patients. Nat. Genet. 1994, 7, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lin, Y.; Yang, X.; Wu, X.; He, X. Long noncoding RNA H19 regulates EZH2 expression by interacting with miR-630 and promotes cell invasion in nasopharyngeal carcinoma. Biochem. Biophys. Res. Commun. 2016, 473, 913–919. [Google Scholar] [CrossRef] [PubMed]
- Imig, J.; Brunschweiger, A.; Brummer, A.; Guennewig, B.; Mittal, N.; Kishore, S.; Tsikrika, P.; Gerber, A.P.; Zavolan, M.; Hall, J. miR-CLIP capture of a miRNA targetome uncovers a lincRNA H19-miR-106a interaction. Nat. Chem. Biol. 2015, 11, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Kallen, A.N.; Zhou, X.B.; Xu, J.; Qiao, C.; Ma, J.; Yan, L.; Lu, L.; Liu, C.; Yi, J.S.; Zhang, H.; et al. The imprinted H19 lncRNA antagonizes Let-7 microRNAs. Mol. Cell 2013, 52, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Keniry, A.; Oxley, D.; Monnier, P.; Kyba, M.; Dandolo, L.; Smits, G.; Reik, W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol. 2012, 14, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Ohtsuka, M.; Ling, H.; Ivan, C.; Pichler, M.; Matsushita, D.; Goblirsch, M.; Stiegelbauer, V.; Shigeyasu, K.; Zhang, X.; Chen, M.; et al. H19 Noncoding RNA, an Independent Prognostic Factor, Regulates Essential Rb-E2F and CDK8-β-Catenin Signaling in Colorectal Cancer. EBioMedicine 2016, 13, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Firestein, R.; Bass, A.J.; Kim, S.Y.; Dunn, I.F.; Silver, S.J.; Guney, I.; Freed, E.; Ligon, A.H.; Vena, N.; Ogino, S.; et al. CDK8 is a colorectal cancer oncogene that regulates β-catenin activity. Nature 2008, 455, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, A.; Goldberg, M.S.; Cumberland, L.K.; Ratnakumar, K.; Segura, M.F.; Emanuel, P.O.; Menendez, S.; Vardabasso, C.; Leroy, G.; Vidal, C.I.; et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature 2010, 468, 1105–1109. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, X.; Liu, Y.; Yuan, J.; Yang, F.; Huang, J.; Meng, Q.; Zhou, C.; Liu, F.; Ma, J.; et al. Long noncoding RNA H19 inhibits the proliferation of fetal liver cells and the Wnt signaling pathway. FEBS Lett. 2016, 590, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Barsyte-Lovejoy, D.; Lau, S.K.; Boutros, P.C.; Khosravi, F.; Jurisica, I.; Andrulis, I.L.; Tsao, M.S.; Penn, L.Z. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006, 66, 5330–5337. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Yu, X.; Ni, B.; Chen, D.; Yang, Z.; Huang, J.; Wang, J.; Chen, D.; Wang, L. Overexpression of long non-coding RNA-CTD903 inhibits colorectal cancer invasion and migration by repressing Wnt/β-catenin signaling and predicts favorable prognosis. Int. J. Oncol. 2016, 48, 2675–2685. [Google Scholar] [CrossRef] [PubMed]
- Taniue, K.; Kurimoto, A.; Takeda, Y.; Nagashima, T.; Okada-Hatakeyama, M.; Katou, Y.; Shirahige, K.; Akiyama, T. ASBEL-TCF3 complex is required for the tumorigenicity of colorectal cancer cells. Proc. Natl. Acad. Sci. USA 2016, 113, 12739–12744. [Google Scholar]
- Kawasaki, Y.; Komiya, M.; Matsumura, K.; Negishi, L.; Suda, S.; Okuno, M.; Yokota, N.; Osada, T.; Nagashima, T.; Hiyoshi, M.; et al. MYU, a Target lncRNA for Wnt/c-Myc Signaling, Mediates Induction of CDK6 to Promote Cell Cycle Progression. Cell Rep. 2016, 16, 2554–2564. [Google Scholar] [CrossRef] [PubMed]
- Ling, H.; Girnita, L.; Buda, O.; Calin, G.A. Non-coding RNAs: The cancer genome dark matter that matters! Clin. Chem. Lab. Med. 2017, 55, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Rittenhouse, H.; Blase, A.; Shamel, B.; Schalken, J.; Groskopf, J. The long and winding road to FDA approval of a novel prostate cancer test: Our story. Clin. Chem. 2013, 59, 32–34. [Google Scholar] [CrossRef] [PubMed]
- McCleland, M.L.; Mesh, K.; Lorenzana, E.; Chopra, V.S.; Segal, E.; Watanabe, C.; Haley, B.; Mayba, O.; Yaylaoglu, M.; Gnad, F.; et al. CCAT1 is an enhancer-templated RNA that predicts BET sensitivity in colorectal cancer. J. Clin. Investig. 2016, 126, 639–652. [Google Scholar] [CrossRef] [PubMed]
- Nan, H.; Morikawa, T.; Suuriniemi, M.; Imamura, Y.; Werner, L.; Kuchiba, A.; Yamauchi, M.; Hunter, D.J.; Kraft, P.; Giovannucci, E.L.; et al. Aspirin use, 8q24 single nucleotide polymorphism rs6983267, and colorectal cancer according to CTNNB1 alterations. J. Natl. Cancer Inst. 2013, 105, 1852–1861. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yang, L.; Zhong, T.; Mueller, M.; Men, Y.; Zhang, N.; Xie, J.; Giang, K.; Chung, H.; Sun, X.; et al. H19 lncRNA alters DNA methylation genome wide by regulating S-adenosylhomocysteine hydrolase. Nat. Commun. 2015, 6, 10221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dang, C.V.; Reddy, E.P.; Shokat, K.M.; Soucek, L. Drugging the ‘undruggable’ cancer targets. Nat. Rev. Cancer 2017, 17, 502–508. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.B.; Johnston, R.L.; Inostroza-Ponta, M.; Fox, A.H.; Fortini, E.; Moscato, P.; Dinger, M.E.; Mattick, J.S. Genome-wide analysis of long noncoding RNA stability. Genome Res. 2012, 22, 885–898. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ling, H.; Fabbri, M.; Calin, G.A. MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat. Rev. Drug Discov. 2013, 12, 847–865. [Google Scholar] [CrossRef] [PubMed]
- Geary, R.S.; Norris, D.; Yu, R.; Bennett, C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015, 87, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Prakash, T.P.; Graham, M.J.; Yu, J.; Carty, R.; Low, A.; Chappell, A.; Schmidt, K.; Zhao, C.; Aghajan, M.; Murray, H.F.; et al. Targeted delivery of antisense oligonucleotides to hepatocytes using triantennary N-acetyl galactosamine improves potency 10-fold in mice. Nucleic Acids Res. 2014, 42, 8796–8807. [Google Scholar] [CrossRef] [PubMed]
- Crooke, S.T.; Wang, S.; Vickers, T.A.; Shen, W.; Liang, X.H. Cellular uptake and trafficking of antisense oligonucleotides. Nat. Biotechnol. 2017, 35, 230–237. [Google Scholar] [CrossRef] [PubMed]

| LncRNA | Loci | Length | Indentification Method | Mechanism Related to Wnt | Clinical Relevance |
|---|---|---|---|---|---|
| 8q24 region | |||||
| CCAT1-S (CARLo-5) | 8q24.21 | 2628 nt | RACE qRT-PCR | Transcription of CCAT1-S is influenced by the allele status of the single nucleotide polymorphisms (SNP) rs6983267 via a long-range interaction of CCAT1-S promoter with rs6938267-containing region. | Promotes CRC growth and invasion; Increased expression correlates with poor prognosis. |
| CCAT1-L | 8q24.21 | 5200 nt | RNA-seq qRT-PCR Northern blot RACE | Interacts with CTCF to faciliate chromatin looping connecting MYC enhancer and promoter, resulting in MYC transcription. | |
| CCAT2 | 8q24.21 | 340 nt | qRT-PCR Northern blot RACE | Interacts with TCF7L2 to promote MYC and other Wnt target gene transcription. Spans the SNP rs6983267 alleles that responds differentially to Wnt signaling. | Promotes CRC growth and metastasis; Increased expression correlates with poor prognosis. |
| CASC11 (CARLo-7) | 8q24.21 | 872 nt | qRT-PCR | Interacts with heterogeneous ribonucleoprotein K (hnRNP-K) to protects β-catenin from degradation, and consequently activates Wnt signaling. MYC binds to the promoter of CASC11 to activate its transcription. | Promotes CRC growth and invasion; Increased expression correlates with CRC size, invasion, and lymph metastasis. |
| PVT1 | 8q24.21 | 1957 nt | RACE Northern blot qRT-PCR | Interacts with MYC protein to prevent MYC phosphorylation and degradation. | Promotes CRC growth. Increased expression correlates with poor prognosis. |
| PCAT1 | 8q24.21 | 1992 nt | qRT-PCR, RNA-seq | Increases MYC expression. | Promotes CRC growth. |
| CRC stem cell | |||||
| Lnc34a | 1p36.22? | 693 nt | qRT-PCR RACE | Interacts with Dnmt3a, HDAC1, and PHB2E to epigenetically silences miR-34a expression, resulting in CRC stem cell asymmetric division. | Enriched in CRC stem cells, and upregulated in late-stage CRCs. |
| RBM5-AS1 | 3p21.31 | 1386 nt | lncRNA array RNA-Seq qRT-PCR | Interacts with β-catenin, and promotes the transcriptional activity of β-catenin/TCF7L2 complex. | |
| WiNTRLINC1 | 11p15.5 | 4117 nt | qRT-PCR Northern blot | Interacts with TCF7L2/β-catenin to form chromatin loop and activate ASCL2 transcription. | Increased expression correlates with metastatic potential and poor prognosis. |
| Others | |||||
| H19 | 11q15.5 | 6295 nt | RACE cloning Northern blot qRT-PCR | Interacts with macroH2A to derepress transcription of CDK8, which positively regulates β-catenin activity. Interacts with hnRNP U to repress Wnt gene transcription. Interacts with EZH2 to repress NKD1 transcription, resulting in Wnt activation. Antagonizes the inhibition of let7 on MYC, which regulates H19 transcription. | Increased expression correlates with poor prognosis independent of other factors. |
| CCAL | Chr3 | 1933 nt | Microarray RACE qRT-PCR | Interacts with and degrades AP-2α, a negative regulator of Wnt activity, resulting in increased MDR1 transcription. | Increased expression correlates with poor prognosis and poor response to adjuvant chemotherapy |
| CTD903 | 14q11.2 | 903 nt | Microarray qRT-PCR | Inhibits Wnt signaling and EMT by unknown mechanisms. | Increased expression correlates with favorable prognosis. |
| ASBEL | 21q21.1 | 2000 nt | qRT-PCR Northern blot | Interacts with TCF3 to repress ATF3 transcription. | |
| MYU | 16q24.3 | 6310 nt | RNA-seq | Upregulated by MYC. Interacts with hnRNP-K to stabilize CDK6 mRNA. |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, P.; Pichler, M.; Chen, M.; Calin, G.A.; Ling, H. To Wnt or Lose: The Missing Non-Coding Linc in Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 2003. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18092003
Shen P, Pichler M, Chen M, Calin GA, Ling H. To Wnt or Lose: The Missing Non-Coding Linc in Colorectal Cancer. International Journal of Molecular Sciences. 2017; 18(9):2003. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18092003
Chicago/Turabian StyleShen, Peng, Martin Pichler, Meng Chen, George A. Calin, and Hui Ling. 2017. "To Wnt or Lose: The Missing Non-Coding Linc in Colorectal Cancer" International Journal of Molecular Sciences 18, no. 9: 2003. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms18092003





