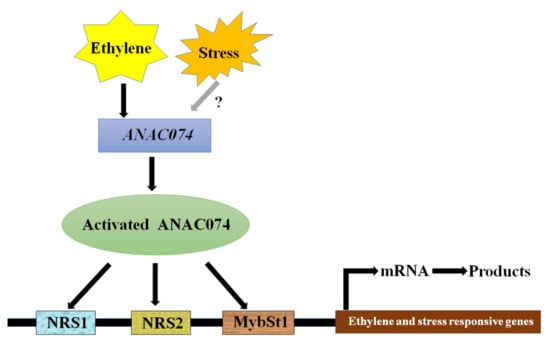
Transcription Factor ANAC074 Binds to NRS1, NRS2, or MybSt1 Element in Addition to the NACRS to Regulate Gene Expression
Abstract
:1. Introduction
2. Results and Discussion
2.1. Identification of the DNA Motifs Bound by ANAC074
2.2. Analysis of the Unknown Elements Recognized by ANAC074
2.3. Binding of Two Novel Motifs and MybSt1 Motif to ANAC074
2.4. ANAC074 Can Bind the Promoters of AT4G17500, AT1G74930 and AT4G11280 via the NRS1, NRS2 or MybSt1 Element
2.5. Binding of ANAC074 to NRS1, NRS2 and MybSt1 Actually Occurs in Plants
2.6. Comparison of the Binding Abilities of ANAC074 to NRS1, NRS2, MybSt1, and NACRS
2.7. Binding of NRS1, NRS2, and MybSt1 to NAC (NAM, ATAF1/2, and CUC2) Family Members
3. Materials and Methods
3.1. TF-Centered Y1H Analysis
3.2. Identification of the Insertion Sequences
3.3. Assay of ANAC074 Binding to the Two Novel Motifs and the MybSt1 Motif
3.4. Analysis of the Binding of ANAC074 to the Truncated Promoters
3.5. ChIP Analysis
3.6. Comparison of ANAC074 Binding with the Four cis-Acting Elements
3.7. Analysis of the Interaction of the Other Ten Arabidopsis NAC Family Members with ANAC074-Binding Elements
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Walhout, A.J. Unraveling transcription regulatory networks by protein-DNA and protein-protein interaction mapping. Genome Res. 2006, 16, 1445–1454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis Transcription Factors: Genome-Wide Comparative Analysis among Eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Nuruzzaman, M.; Manimekalai, R.; Sharoni, A.M.; Satoh, K.; Kondoh, H.; Ooka, H.; Kikuchi, S. Genome-wide analysis of NAC transcription factor family in rice. Gene 2010, 465, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Souer, E.; van Houwelingen, A.; Kloos, D.; Mol, J.; Koes, R. The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 1996, 85, 159–170. [Google Scholar] [CrossRef]
- Xie, Q.; Frugis, G.; Colgan, D.; Chua, N.H. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000, 14, 3024–3036. [Google Scholar] [CrossRef] [PubMed]
- Ohashi-Ito, K.; Oda, Y.; Fukuda, H. Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 directly regulates the genes that govern programmed cell death and secondary wall formation during xylem differentiation. Plant Cell 2010, 22, 3461–3473. [Google Scholar] [CrossRef] [PubMed]
- Takasaki, H.; Maruyama, K.; Takahashi, F.; Fujita, M.; Yoshida, T.; Nakashima, K.; Myouga, F.; Toyooka, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. SNAC-As, stress-responsive NAC transcription factors, mediate ABA-inducible leaf senescence. Plant J. 2015, 84, 1114–1123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, K.; Bibi, N.; Gan, S.; Li, F.; Yuan, S.; Ni, M.; Wang, M.; Shen, H.; Wang, X. A novel NAP member GhNAP is involved in leaf senescence in Gossypium hirsutum. J. Exp. Bot. 2015, 66, 4669–4682. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Demura, T.; Ye, Z.H. SND1, a NAC domain transcription factor, is a key regulator of secondary wall synthesis in fibers of Arabidopsis. Plant Cell 2006, 18, 3158–3170. [Google Scholar] [CrossRef] [PubMed]
- Shahnejat-Bushehri, S.; Tarkowska, D.; Sakuraba, Y.; Balazadeh, S. Arabidopsis NAC transcription factor JUB1 regulates GA/BR metabolism and signalling. Nat. Plants 2016, 2, 16013. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Lu, S.; Lv, B.; Zhang, B.; Shen, J.; He, J.; Ming, F. A rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol. 2017, 174, 1747–1763. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Dai, M.; Yao, J.; Xiao, B.; Li, X.; Zhang, Q.; Xiong, L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987–12992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shan, W.; Kuang, J.F.; Lu, W.J.; Chen, J.Y. Banana fruit NAC transcription factor MaNAC1 is a direct target of MaICE1 and involved in cold stress through interacting with MaCBF1. Plant Cell Environ. 2014, 37, 2116–2127. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wang, X.; Ji, L.; Yi, Z.; Fu, C.; Ran, J.; Hu, R.; Zhou, G. Overexpression of a Miscanthus lutarioriparius NAC gene MlNAC5 confers enhanced drought and cold tolerance in Arabidopsis. Plant Cell Rep. 2015, 34, 943–958. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.; Su, L.; Sun, X.; Li, X.; Sun, M.; Karungo, S.K.; Fang, S.; Chu, J.; Li, S.; Xin, H. Expression of Vitis amurensis NAC26 in Arabidopsis enhances drought tolerance by modulating jasmonic acid synthesis. J. Exp. Bot. 2016, 67, 2829–2845. [Google Scholar] [CrossRef] [PubMed]
- Duan, M.; Zhang, R.; Zhu, F.; Zhang, Z.; Gou, L.; Wen, J.; Dong, J.; Wang, T. A Lipid-anchored NAC transcription factor is translocated into the nucleus and activates glyoxalase I expression during drought stress. Plant Cell 2017, 29, 1748–1772. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xia, Y.; Lin, S.; Wang, Y.; Guo, B.; Song, X.; Ding, S.; Zheng, L.; Feng, R.; Chen, S.; et al. Osa-miR164a targets OsNAC60 and negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Plant J. 2018. [Google Scholar] [CrossRef] [PubMed]
- Tran, L.S.; Nakashima, K.; Sakuma, Y.; Simpson, S.D.; Fujita, Y.; Maruyama, K.; Fujita, M.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 2004, 16, 2481–2498. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.Y.; Kim, S.Y.; do Hyeon, Y.; Kim, D.H.; Dong, T.; Park, Y.; Jin, J.B.; Joo, S.H.; Kim, S.K.; Hong, J.C.; et al. The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 2013, 25, 4708–4724. [Google Scholar] [CrossRef] [PubMed]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. DNA-binding specificity and molecular functions of NAC transcription factors. Plant Sci. 2005, 169, 785–797. [Google Scholar] [CrossRef]
- He, L.; Shi, X.X.; Wang, Y.M.; Guo, Y.; Yang, K.J.; Wang, Y.C. Arabidopsis ANAC069 binds to C[A/G]CG[T/G] sequences to negatively regulate salt and osmotic stress tolerance. Plant Mol. Biol. 2016, 93, 369–387. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.Y.; Wang, L.Q.; Nie, X.G.; He, L.; Zang, D.D.; Liu, Y.J.; Zhang, B.; Wang, Y.C. A novel method to identify the DNA motifs recognized by a defined transcription factor. Plant Mol. Biol. 2014, 86, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.Y.; Shi, X.X.; Wang, Z.B.; Gao, C.Q.; Wang, C.; Wang, Y.C. Transcription factor ThWRKY4 binds to a novel WLS motif and a RAV1A element in addition to the W-box to regulate gene expression. Plant Sci. 2017, 261, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Park, B.O.; Yoo, J.H.; Jung, M.S.; Lee, S.M.; Han, H.J.; Kim, K.E.; Kim, S.H.; Lim, C.O.; Yun, D.J.; et al. Identification of a calmodulin-binding NAC protein as a transcriptional repressor in Arabidopsis. J. Biol. Chem. 2007, 282, 36292–36302. [Google Scholar] [CrossRef] [PubMed]
- Baranowskij, N.; Frohberg, C.; Prat, S.; Willmitzer, L. A novel DNA binding protein with homology to Myb oncoproteins containing only one repeat can function as a transcriptional activator. EMBO J. 1994, 13, 5383–5392. [Google Scholar] [CrossRef] [PubMed]
- Czemmel, S.; Stracke, R.; Weisshaar, B.; Cordon, N.; Harris, N.N.; Walker, A.R.; Robinson, S.P.; Bogs, J. The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol. 2009, 151, 1513–1530. [Google Scholar] [CrossRef] [PubMed]
- Muller, D.R.; Schmitz, G.; Theres, K. Blind homologous R2R3 Myb genes control the pattern of lateral meristem initiation in Arabidopsis. Plant Cell 2006, 18, 586–597. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.; Jiang, T.; Xu, L.; Liu, H.; Mao, H.; Wang, X.; Jiao, B.; Duan, Y.; Wang, Q.; Dong, Q.; et al. A salt-stress-regulator from the Poplar R2R3 MYB family integrates the regulation of lateral root emergence and ABA signaling to mediate salt stress tolerance in Arabidopsis. Plant Physiol. Biochem. 2017, 114, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.W.; Qi, M.; Yang, Y. A novel jasmonic acid-inducible rice Myb gene associates with fungal infection and host cell death. Mol. Plant-Microbe Interact. 2001, 14, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ju, H.W.; Chung, M.S.; Huang, P.; Ahn, S.J.; Kim, C.S. The R-R-type MYB-like transcription factor, AtMYBL, is involved in promoting leaf senescence and modulates an abiotic stress response in Arabidopsis. Plant Cell Physiol. 2011, 52, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ng, C.K.Y.; Fan, L.M. MYB transcription factors, active players in abiotic stress signalling. Environ. Exp. Bot. 2015, 114, 80–91. [Google Scholar] [CrossRef]
- Yan, T.; Yoo, D.; Berardini, T.Z.; Mueller, L.A.; Weems, D.C.; Weng, S.; Cherry, J.M.; Rhee, S.Y. PatMatch: A program for finding patterns in peptide and nucleotide sequences. Nucleic Acids Res. 2005, 33, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.M.; Zhao, Q.; Zhao, L.L.; Qiao, Y.; Xie, X.B.; Li, H.F.; Yao, Y.X.; You, C.X.; Hao, Y.J. The cold-induced basic helix-loop-helix transcription factor gene MdCIbHLH1 encodes an ICE-like protein in apple. BMC Plant Biol. 2012, 12, 22. [Google Scholar] [CrossRef] [PubMed]
- Shan, W.; Kuang, J.F.; Chen, L.; Xie, H.; Peng, H.H.; Xiao, Y.Y.; Li, X.P.; Chen, W.X.; He, Q.G.; Chen, J.Y.; et al. Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. J. Exp. Bot. 2012, 63, 5171–5187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, X.J.; Mu, R.L.; Cao, W.H.; Zhang, Z.G.; Zhang, J.S.; Chen, S.Y. AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 2005, 44, 903–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, J.P.; Yao, J.F.; Xu, R.R.; You, C.X.; Wang, X.F.; Hao, Y.J. An apple NAC transcription factor enhances salt stress tolerance by modulating the ethylene response. Physiol. Plant 2018, 164, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.; Liu, C.; Han, L.; Wang, S.; Xue, Z. NAC Transcription factors play an important role in ethylene biosynthesis, reception and signaling of tomato fruit ripening. Mol. Genet. Genom. 2016, 291, 1205–1217. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Chen, Y.; Chen, Y.; Shin, J.H.; Mila, I.; Audran, C.; Zouine, M.; Pirrello, J.; Bouzayen, M. The tomato Ethylene Response Factor Sl-ERF.B3 integrates ethylene and auxin signaling viadirect regulation of Sl-Aux/IAA27. New Phytol. 2018, 219, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.C.; Park, H.J.; Wang, H.Y. Role of Arabidopsis RAP2.4 in regulating light- and ethylene-mediated developmental processes and drought stress tolerance. Mol. Plant 2008, 1, 42–57. [Google Scholar] [CrossRef] [PubMed]
- Guan, R.; Su, J.; Meng, X.; Li, S.; Liu, Y.; Xu, J.; Zhang, S. Multilayered Regulation of ethylene Induction plays a positive role in Arabidopsis resistance against Pseudomonas syringae. Plant Physiol. 2015, 169, 299–312. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Tu, X.; Zhang, J.; Chen, X.; Rao, L. Heat stress-induced BBX18 negatively regulates the thermotolerance in Arabidopsis. Mol. Biol. Rep. 2013, 40, 2679–2688. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Shibuya, H.; Ishikawa, A. SOBIR1 contributes to non-host resistance to Magnaporthe Oryzae in Arabidopsis. Biosci. Biotechnol. Biochem. 2016, 80, 1577–1579. [Google Scholar] [CrossRef] [PubMed]
- Liebrand, T.W.; van den Berg, G.C.; Zhang, Z.; Smit, P.; Cordewener, J.H.; America, A.H.; Sklenar, J.; Jones, A.M.; Tameling, W.I.; Robatzek, S.; et al. Receptor-like kinase SOBIR1/EVR Interacts with receptor-like proteins in plant immunity against fungal infection. Proc. Natl. Acad. Sci. USA 2013, 110, 10010–10015. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.K.; Kjaersgaard, T.; Nielsen, M.M.; Galberg, P.; Petersen, K.; O’Shea, C.; Skriver, K. The Arabidopsis thaliana NAC transcription factor family: Structure–function relationships and determinants of ANAC019 stress signaling. Biochem. J. 2010, 426, 183–196. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Betaglucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef] [PubMed]
- Haring, M.; Offermann, S.; Danker, T.; Horst, I.; Peterhansel, C.; Stam, M. Chromatin immunoprecipitation: Optimization, quantitative analysis and data normalization. Plant Methods 2007, 3, 11. [Google Scholar] [CrossRef] [PubMed]






| Clone Number | Random DNA Insertion Sequence (5’–3’) Underlined Together with the Two Sides of Flanking Sequences | Motif Prediction |
|---|---|---|
| 1 | CCCCTTCACGCGGG | Core sequence of NACRS: “CACG” |
| 2 | CCCAGGATAACGGG | MybSt1-binding element: “GGATA” |
| 3 | CCCTTAGTTTCGGG | LTRE-1: “CCGAAA” |
| 4 | CCCTGAATCTCGGG | ARR1AT: “NGATT” |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, L.; Xu, J.; Wang, Y.; Yang, K. Transcription Factor ANAC074 Binds to NRS1, NRS2, or MybSt1 Element in Addition to the NACRS to Regulate Gene Expression. Int. J. Mol. Sci. 2018, 19, 3271. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19103271
He L, Xu J, Wang Y, Yang K. Transcription Factor ANAC074 Binds to NRS1, NRS2, or MybSt1 Element in Addition to the NACRS to Regulate Gene Expression. International Journal of Molecular Sciences. 2018; 19(10):3271. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19103271
Chicago/Turabian StyleHe, Lin, Jingyu Xu, Yucheng Wang, and Kejun Yang. 2018. "Transcription Factor ANAC074 Binds to NRS1, NRS2, or MybSt1 Element in Addition to the NACRS to Regulate Gene Expression" International Journal of Molecular Sciences 19, no. 10: 3271. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19103271