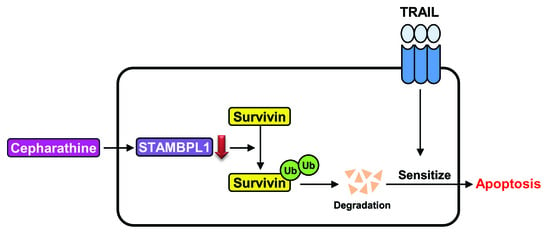
Cepharanthine Enhances TRAIL-Mediated Apoptosis Through STAMBPL1-Mediated Downregulation of Survivin Expression in Renal Carcinoma Cells
Abstract
:1. Introduction
2. Results
2.1. CEP Sensitizes TRAIL-Induced Apoptosis in Human Renal Carcinoma Caki Cells
2.2. CEP Did Not Increase DR5 Expression on the Cell Surface
2.3. CEP Induced Downregulation of c-FLIP Protein Expression
2.4. Downregulation of Survivin by CEP is Critical to Sensitization of TRAIL-Mediated Apoptosis
2.5. Combined CEP and TRAIL Treatment Enhances Apoptosis in Other Cancer Cells, but Not Normal Cells
3. Discussion
4. Methods and Materials
4.1. Cells and Cell Culture Materials
4.2. Flow Cytometry Analysis and Western Blot Analysis
4.3. Asp–Glu–Val–Asp-ase (DEVDase) Activity Assay
4.4. Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
4.5. Detection of DR5 on Cell Surface
4.6. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| TRAIL | TNF-related apoptosis-inducing ligand |
| CEP | Cepharanthine |
| DR5 | death receptor 5 |
| STAMBPL1 | STAM-binding protein-like 1 |
| CHX | Cycloheximide |
References
- Hofmann, H.S.; Neef, H.; Krohe, K.; Andreev, P.; Silber, R.E. Prognostic factors and survival after pulmonary resection of metastatic renal cell carcinoma. Eur. Urol. 2005, 48, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Vitale, M.G.; Carteni, G. Recent developments in second and third line therapy of metastatic renal cell carcinoma. Expert Rev. Anticancer Ther. 2016, 16, 469–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patil, S.; Ishill, N.; Deluca, J.; Motzer, R.J. Stage migration and increasing proportion of favorable-prognosis metastatic renal cell carcinoma patients: Implications for clinical trial design and interpretation. Cancer 2010, 116, 347–354. [Google Scholar] [CrossRef] [PubMed]
- Kassouf, W.; Aprikian, A.G.; Laplante, M.; Tanguay, S. Natural history of renal masses followed expectantly. J. Urol. 2004, 171, 111–113. [Google Scholar] [CrossRef] [PubMed]
- Coppin, C.; Le, L.; Porzsolt, F.; Wilt, T. Targeted therapy for advanced renal cell carcinoma. Cochrane Database Syst. Rev. 2008, 2, CD006017. [Google Scholar] [CrossRef] [PubMed]
- Albiges, L.; Oudard, S.; Negrier, S.; Caty, A.; Gravis, G.; Joly, F.; Duclos, B.; Geoffrois, L.; Rolland, F.; Guillot, A.; et al. Complete remission with tyrosine kinase inhibitors in renal cell carcinoma. J. Clin. Oncol. 2012, 30, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Posadas, E.M.; Limvorasak, S.; Figlin, R.A. Targeted therapies for renal cell carcinoma. Nat. Rev. Nephrol. 2017, 13, 496–511. [Google Scholar] [CrossRef] [PubMed]
- French, L.E.; Tschopp, J. The TRAIL to selective tumor death. Nat. Med. 1999, 5, 146–147. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; El-Deiry, W.S. TRAIL and apoptosis induction by TNF-family death receptors. Oncogene 2003, 22, 8628–8633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, G.; Ni, J.; Wei, Y.F.; Yu, G.; Gentz, R.; Dixit, V.M. An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science 1997, 277, 815–818. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, J.P.; Marsters, S.A.; Pitti, R.M.; Gurney, A.; Skubatch, M.; Baldwin, D.; Ramakrishnan, L.; Gray, C.L.; Baker, K.; Wood, W.I.; et al. Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science 1997, 277, 818–821. [Google Scholar] [CrossRef] [PubMed]
- Gulcin, I.; Elias, R.; Gepdiremen, A.; Chea, A.; Topal, F. Antioxidant activity of bisbenzylisoquinoline alkaloids from Stephania rotunda: Cepharanthine and fangchinoline. J. Enzyme Inhib. Med. Chem. 2010, 25, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Rogosnitzky, M.; Danks, R. Therapeutic potential of the biscoclaurine alkaloid, cepharanthine, for a range of clinical conditions. Pharmacol. Rep. 2011, 63, 337–347. [Google Scholar] [CrossRef]
- Furusawa, S.; Wu, J.; Fujimura, T.; Nakano, S.; Nemoto, S.; Takayanagi, M.; Sasaki, K.; Takayanagi, Y. Cepharanthine inhibits proliferation of cancer cells by inducing apoptosis. Methods Find Exp. Clin. Pharmacol. 1998, 20, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Kikukawa, Y.; Okuno, Y.; Tatetsu, H.; Nakamura, M.; Harada, N.; Ueno, S.; Kamizaki, Y.; Mitsuya, H.; Hata, H. Induction of cell cycle arrest and apoptosis in myeloma cells by cepharanthine, a biscoclaurine alkaloid. Int. J. Oncol. 2008, 33, 807–814. [Google Scholar] [PubMed]
- Uthaisar, K.; Seubwai, W.; Srikoon, P.; Vaeteewoottacharn, K.; Sawanyawisuth, K.; Okada, S.; Wongkham, S. Cepharanthine suppresses metastatic potential of human cholangiocarcinoma cell lines. Asian Pac. J. Cancer Prev. 2012, 13, 149–154. [Google Scholar] [PubMed]
- Biswas, K.K.; Tancharoen, S.; Sarker, K.P.; Kawahara, K.; Hashiguchi, T.; Maruyama, I. Cepharanthine triggers apoptosis in a human hepatocellular carcinoma cell line (HuH-7) through the activation of JNK1/2 and the downregulation of Akt. FEBS Lett. 2006, 580, 703–710. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Suzuki, H.; Zhou, Y.W.; Liu, W.; Yoshihara, M.; Kato, M.; Akhand, A.A.; Hayakawa, A.; Takeuchi, K.; Hossain, K.; et al. Cepharanthine activates caspases and induces apoptosis in Jurkat and K562 human leukemia cell lines. J. Cell Biochem. 2001, 82, 200–214. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Huang, C.; Yang, Y.L.; Ding, Y.; Ou-Yang, H.Q.; Zhang, Y.Y.; Xu, M. Inhibition of the STAT3 signaling pathway is involved in the antitumor activity of cepharanthine in SaOS2 cells. Acta Pharmacol. Sin. 2012, 33, 101–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seubwai, W.; Vaeteewoottacharn, K.; Hiyoshi, M.; Suzu, S.; Puapairoj, A.; Wongkham, C.; Okada, S.; Wongkham, S. Cepharanthine exerts antitumor activity on cholangiocarcinoma by inhibiting NF-kappaB. Cancer Sci. 2010, 101, 1590–1595. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Turcu, F.E.; Ventii, K.H.; Wilkinson, K.D. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 2009, 78, 363–397. [Google Scholar] [CrossRef] [PubMed]
- Hua, P.; Sun, M.; Zhang, G.; Zhang, Y.; Tian, X.; Li, X.; Cui, R.; Zhang, X. Cepharanthine induces apoptosis through reactive oxygen species and mitochondrial dysfunction in human non-small-cell lung cancer cells. Biochem. Biophys. Res. Commun. 2015, 460, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Furusawa, S.; Wu, J. The effects of biscoclaurine alkaloid cepharanthine on mammalian cells: Implications for cancer, shock, and inflammatory diseases. Life Sci. 2007, 80, 1073–1079. [Google Scholar] [CrossRef] [PubMed]
- Rattanawong, A.; Payon, V.; Limpanasittikul, W.; Boonkrai, C.; Mutirangura, A.; Wonganan, P. Cepharanthine exhibits a potent anticancer activity in p53-mutated colorectal cancer cells through upregulation of p21Waf1/Cip1. Oncol. Rep. 2018, 39, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.G.; Yu, F.; Yao, Q.; Chen, J.H.; Wang, L. The role of survivin in diagnosis, prognosis and treatment of breast cancer. J. Thorac. Dis. 2010, 2, 100–110. [Google Scholar] [PubMed]
- Shoeneman, J.K.; Ehrhart, E.J., 3rd; Eickhoff, J.C.; Charles, J.B.; Powers, B.E.; Thamm, D.H. Expression and function of survivin in canine osteosarcoma. Cancer Res. 2012, 72, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Boidot, R.; Vegran, F.; Lizard-Nacol, S. Transcriptional regulation of the survivin gene. Mol. Biol. Rep. 2014, 41, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yang, F.; Li, X.; Gong, Z.J.; Wang, L.W. Long noncoding RNA LNC473 inhibits the ubiquitination of survivin via association with USP9X and enhances cell proliferation and invasion in hepatocellular carcinoma cells. Biochem. Biophys. Res. Commun. 2018, 499, 702–710. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lear, T.; Iannone, O.; Shiva, S.; Corey, C.; Rajbhandari, S.; Jerome, J.; Chen, B.B.; Mallampalli, R.K. The Proapoptotic F-box Protein Fbxl7 Regulates Mitochondrial Function by Mediating the Ubiquitylation and Proteasomal Degradation of Survivin. J. Biol. Chem. 2015, 290, 11843–11852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Skaug, B.; Jiang, X.; Chen, Z.J. The role of ubiquitin in NF-kappaB regulatory pathways. Annu. Rev. Biochem. 2009, 78, 769–796. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, K.D. DUBs at a glance. J. Cell Sci. 2009, 122, 2325–2329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavorgna, A.; Harhaj, E.W. An RNA interference screen identifies the Deubiquitinase STAMBPL1 as a critical regulator of human T-cell leukemia virus type 1 tax nuclear export and NF-kappaB activation. J. Virol. 2012, 86, 3357–3369. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.H.; Altieri, D.C. Regulation of survivin stability by the aryl hydrocarbon receptor-interacting protein. J. Biol. Chem. 2006, 281, 24721–24727. [Google Scholar] [CrossRef] [PubMed]
- Arora, V.; Cheung, H.H.; Plenchette, S.; Micali, O.C.; Liston, P.; Korneluk, R.G. Degradation of survivin by the X-linked inhibitor of apoptosis (XIAP)-XAF1 complex. J. Biol. Chem. 2007, 282, 26202–26209. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Fueger, P.T.; Wang, Z. Depletion of PAK1 enhances ubiquitin-mediated survivin degradation in pancreatic beta-cells. Islets 2013, 5, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Seo, B.R.; Min, K.J.; Woo, S.M.; Choe, M.; Choi, K.S.; Lee, Y.K.; Yoon, G.; Kwon, T.K. Inhibition of Cathepsin S Induces Mitochondrial ROS That Sensitizes TRAIL-Mediated Apoptosis Through p53-Mediated Downregulation of Bcl-2 and c-FLIP. Antioxid. Redox. Signal. 2017, 27, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Jo, Y.; Shin, D.Y. Repression of the F-box protein Skp2 is essential for actin damage-induced tetraploid G1 arrest. BMB Rep. 2017, 50, 379–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, Y.S.; Kwon, Y.J.; Chun, Y.J. CYP1B1 Activates Wnt/beta-Catenin Signaling through Suppression of Herc5-Mediated ISGylation for Protein Degradation on beta-Catenin in HeLa Cells. Toxicol. Res. 2017, 33, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.U.; Min, K.J.; Woo, S.M.; Kwon, T.K. Z-FL-COCHO, a cathepsin S inhibitor, enhances oxaliplatin-mediated apoptosis through the induction of endoplasmic reticulum stress. Exp. Mol. Med. 2018, 50, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.M.; Min, K.J.; Seo, S.U.; Kim, S.; Park, J.W.; Song, D.K.; Lee, H.S.; Kim, S.H.; Kwon, T.K. Up-regulation of 5-lipoxygenase by inhibition of cathepsin G enhances TRAIL-induced apoptosis through down-regulation of survivin. Oncotarget 2017, 8, 106672–106684. [Google Scholar] [CrossRef] [PubMed]





© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahriyar, S.A.; Woo, S.M.; Seo, S.U.; Min, K.-j.; Kwon, T.K. Cepharanthine Enhances TRAIL-Mediated Apoptosis Through STAMBPL1-Mediated Downregulation of Survivin Expression in Renal Carcinoma Cells. Int. J. Mol. Sci. 2018, 19, 3280. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19103280
Shahriyar SA, Woo SM, Seo SU, Min K-j, Kwon TK. Cepharanthine Enhances TRAIL-Mediated Apoptosis Through STAMBPL1-Mediated Downregulation of Survivin Expression in Renal Carcinoma Cells. International Journal of Molecular Sciences. 2018; 19(10):3280. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19103280
Chicago/Turabian StyleShahriyar, Sk Abrar, Seon Min Woo, Seung Un Seo, Kyoung-jin Min, and Taeg Kyu Kwon. 2018. "Cepharanthine Enhances TRAIL-Mediated Apoptosis Through STAMBPL1-Mediated Downregulation of Survivin Expression in Renal Carcinoma Cells" International Journal of Molecular Sciences 19, no. 10: 3280. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19103280