Fingerprinting Acute Digestive Diseases by Untargeted NMR Based Metabolomics
Abstract
:1. Introduction
2. Results
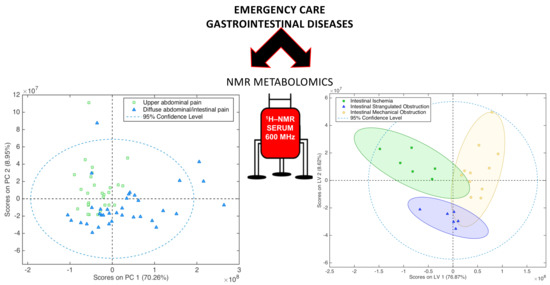
2.1. Diseases Discrimination According to Their Symptoms
2.2. Biliary/Pancreatic Diseases
2.3. Intestinal Diseases
3. Discussion
4. Materials and Methods
4.1. Patients Recruitment for the Study
4.2. Blood Collection and Preparation
4.3. NMR Spectra Processing and Statistical Analysis Methods
4.4. Data Deposition
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| LC-MS | Liquid Chromatography–Mass Spectrometry |
| NMR | Nuclear Magnetic Resonance |
| NOESY | Nuclear Overhauser Effect Spectroscopy |
| CPMG | Carr-Purcell-Meiboom-Gill |
| UAP | Upper Abdominal Pain |
| DAIP | Diffuse Abdominal/Intestinal Pain |
| HMDB | Human Metabolome Database |
| LV | Latent Variable |
| OPLS-DA | Orthogonal Partial Least Square–Discriminant Analysis |
| MVDA | Multivariate Discriminant Analysis |
| VIP | Variable Importance in Projection |
| PCA | Principal Component Analysis |
| PE | Phosphatidylethanolamine |
| PC | Phosphatidylcholine |
References
- Whitcomb, D.C. What is personalized medicine and what should it replace? Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 418–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milne, C.-P.; Cohen, J.P.; Chakravarthy, R. Market watch: Where is personalized medicine in industry heading? Nat. Rev. Drug Discov. 2015, 12, 812–813. [Google Scholar] [CrossRef] [PubMed]
- Rattray, N.J.W.; Hamrang, Z.; Trivedi, D.K.; Goodacre, R.; Fowler, S.J. Taking your breath away: Metabolomics breathes life in to personalized medicine. Trends Biotechnol. 2014, 32, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Psychogios, N.; Hau, D.D.; Peng, J.; Guo, A.C.; Mandal, R.; Bouatra, S.; Sinelnikov, I.; Krishnamurthy, R.; Eisner, R.; Gautam, B.; et al. The human serum metabolome. PLoS ONE 2011, 6, e16957. [Google Scholar] [CrossRef] [PubMed]
- Takis, P.G.; Schäfer, H.; Spraul, M.; Luchinat, C. Deconvoluting interrelationships between concentrations and chemical shifts in urine provides a powerful analysis tool. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Lindon, J.C.; Nicholson, J.K. Spectroscopic and Statistical Techniques for Information Recovery in Metabonomics and Metabolomics. Annu. Rev. Anal. Chem. 2008, 1, 45–69. [Google Scholar] [CrossRef] [PubMed]
- Sands, B.E. Biomarkers of Inflammation in Inflammatory Bowel Disease. Gastroenterology 2015, 149, 1275–1285. [Google Scholar] [CrossRef] [PubMed]
- Bjerrum, J.T.; Nielsen, O.H.; Hao, F.; Tang, H.; Nicholson, J.K.; Wang, Y.; Olsen, J. Metabonomics in ulcerative colitis: Diagnostics, biomarker identification, and insight into the pathophysiology. J. Proteome Res. 2010, 9, 954–962. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.R.; McPhail, M.J.; Shariff, M.I.; Keun, H.C.; Taylor-Robinson, S.D. Biofluid metabolomics using 1H NMR spectroscopy: The road to biomarker discovery in gastroenterology and hepatology. Expert Rev. Gastroenterol. Hepatol. 2012, 6, 239–251. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, O.J.; O’Neill, S.; Maher, M.M. Imaging of Biliary Tract Disease. Am. J. Roentgenol. 2011, 197, W551–W558. [Google Scholar] [CrossRef] [PubMed]
- Safrany, L. Endoscopic treatment of biliary-tract diseases: An International Study. Lancet 1978, 312, 983–985. [Google Scholar] [CrossRef]
- Gallegos, N.C.; Dawson, J.; Jarvis, M.; Hobsley, M. Risk of strangulation in groin hernias. Br. J. Surg. 1991, 78, 1171–1173. [Google Scholar] [CrossRef] [PubMed]
- Lieberman, J.M.; Sacchettini, J.; Marks, C.; Marks, W.H. Human intestinal fatty acid binding protein: Report of an assay with studies in normal volunteers and intestinal ischemia. Surgery 1997, 121, 335–342. [Google Scholar] [CrossRef]
- Rychter, J.; Clavé, P. Intestinal inflammation in postoperative ileus: Pathogenesis and therapeutic targets. Gut 2013, 62, 1534–1535. [Google Scholar] [CrossRef] [PubMed]
- Thuijls, G.; van Wijck, K.; Grootjans, J.; Derikx, J.P.M.; van Bijnen, A.A.; Heineman, E.; Dejong, C.H.C.; Buurman, W.A.; Poeze, M. Early Diagnosis of Intestinal Ischemia Using Urinary and Plasma Fatty Acid Binding Proteins. Ann. Surg. 2011, 253, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Turano, P. Colorectal cancer: The potential of metabolic fingerprinting. Expert Rev. Gastroenterol. Hepatol. 2014, 8, 847–849. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.-L.; Cen, Y.-Y.; Li, S.-M.; Li, W.-M.; Lu, Q.-P.; Xu, P.-Y. Accuracy of the serum intestinal fatty-acid-binding protein for diagnosis of acute intestinal ischemia: A meta-analysis. Sci. Rep. 2016, 6, 34371. [Google Scholar] [CrossRef] [PubMed]
- Blankenberg, F.; Wirth, R.; Jeffrey, R.B., Jr.; Mindelzun, R.; Francis, I. Computed tomography as an adjunct to ultrasound in the diagnosis of acute acalculous cholecystitis. Gastrointest. Radiol. 1991, 16, 149–153. [Google Scholar] [CrossRef] [PubMed]
- Ko, C. Biliary sludge and acute pancreatitis during pregnancy. Nat. Clin. Pract. Gastroenterol. Hepatol. 2006, 3, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Ziessman, H.A. Nuclear Medicine Hepatobiliary Imaging. Clin. Gastroenterol. Hepatol. 2010, 8, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Jeffrey, R.B.; Laing, F.C.; Wong, W.; Callen, P.W. Gangrenous cholecystitis: Diagnosis by ultrasound. Radiology 1983, 148, 219–221. [Google Scholar] [CrossRef] [PubMed]
- Giesbrandt, K.; Young, P. Noninvasive Arterial Imaging: Computed Tomography and Magnetic Resonance Angiography. In Mesenteric Vascular Disease; Oderich, G.S., Ed.; Springer: New York, NY, USA, 2015; pp. 79–94. [Google Scholar]
- Bertini, I.; Cacciatore, S.; Jensen, B.V.; Schou, J.V.; Johansen, J.S.; Kruhøffer, M.; Luchinat, C.; Nielsen, D.L.; Turano, P. Metabolomic NMR Fingerprinting to Identify and Predict Survival of Patients with Metastatic Colorectal Cancer. Cancer Res. 2012, 72, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Sztefko, K.; Panek, J. Serum Free Fatty Acid Concentration in Patients with Acute Pancreatitis. Pancreatology 2001, 1, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Groen, A.K.; Noordam, C.; Drapers, J.A.G.; Egbers, P.; Jansen, P.L.M.; Tytgat, G.N.J. Isolation of a potent cholesterol nucleation-promoting activity from human gallbladder bile: Role in the pathogenesis of gallstone disease. Hepatology 1990, 11, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T.; Nakatomi, Y.; Ishikawa, H.; Hitomi, M.; Matsubara, Y.; Ono, T.; Muto, T. Intestinal fatty acid-binding protein as a sensitive marker of intestinal ischemia. Dig. Dis. Sci. 1992, 37, 1362–1367. [Google Scholar] [CrossRef] [PubMed]
- Murray, M.J.; Gonze, M.D.; Nowak, L.R.; Cobb, C.F. Serum D(−)-lactate levels as an aid to diagnosing acute intestinal ischemia. Am. J. Surg. 2015, 167, 575–578. [Google Scholar] [CrossRef]
- Kittaka, H.; Akimoto, H.; Takeshita, H.; Funaoka, H.; Hazui, H.; Okamoto, M.; Kobata, H.; Ohishi, Y. Usefulness of intestinal fatty acid-binding protein in predicting strangulated small bowel obstruction. PLoS ONE 2014, 9, e99915. [Google Scholar] [CrossRef] [PubMed]
- Agouridis, A.P.; Elisaf, M.; Milionis, H.J. An overview of lipid abnormalities in patients with inflammatory bowel disease. Ann. Gastroenterol. 2011, 24, 181–187. [Google Scholar] [PubMed]
- Evennett, N.; Petrov, M.; Mittal, A.; Windsor, J. Systematic Review and Pooled Estimates for the Diagnostic Accuracy of Serological Markers for Intestinal Ischemia. World J. Surg. 2009, 33, 1374–1383. [Google Scholar] [CrossRef] [PubMed]
- Heimerl, S.; Moehle, C.; Zahn, A.; Boettcher, A.; Stremmel, W.; Langmann, T.; Schmitz, G. Alterations in intestinal fatty acid metabolism in inflammatory bowel disease. Biochim. Biophys. Acta 2006, 1762, 341–350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Preter, V.; Verbeke, K. Metabolomics as a diagnostic tool in gastroenterology. World J. Gastrointest. Pharmacol. Ther. 2013, 4, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Leek, J. Five ways to fix statistics. Nature 2017, 551, 557–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Worley, B.; Powers, R. Multivariate Analysis in Metabolomics. Curr. Metabolomics 2013, 1, 92–107. [Google Scholar] [CrossRef] [PubMed]
- Thomas, E.V. Non-parametric statistical methods for multivariate calibration model selection and comparison. J. Chemom. 2004, 17, 653–659. [Google Scholar] [CrossRef]
- Van der Voet, H. Comparing the predictive accuracy of models using a simple randomization test. Chemom. Intell. Lab. Syst. 1994, 25, 313–323. [Google Scholar] [CrossRef]
- Takis, P.G.; Oraiopoulou, M.-E.; Konidaris, C.; Troganis, A.N. 1H-NMR based metabolomics study for the detection of the human urine metabolic profile effects of Origanum Dictamnus tea ingestion. Food Funct. 2016, 7, 4104–4115. [Google Scholar] [CrossRef] [PubMed]
- Lindon, J.C. Biofluids Studied by NMR. In Encyclopedia of Spectroscopy and Spectrometry, 2nd ed.; Academic Press: Oxford, UK, 2010; pp. 128–141. [Google Scholar]
- Tukiainen, T.; Tynkkynen, T.; Mäkinen, V.P.; Jylänki, P.; Kangas, A.; Hokkanen, J.; Vehtari, A.; Gröhn, O.; Hallikainen, M.; Soininen, H.; et al. Disease. Biochem. Biophys. Res. Commun. 2008, 375, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Tzur, D.; Knox, C.; Eisner, R.; Guo, A.C.; Young, N.; Cheng, D.; Jewell, K.; Arndt, D.; Sawhney, S.; et al. HMDB: The Human Metabolome Database. Nucleic Acids Res. 2007, 35, D521–D526. [Google Scholar] [CrossRef] [PubMed]





| Upper Abdominal Pain Symptom (UAP) | Diffuse Intestinal/Abdominal Pain Symptom (DAIP) | |
|---|---|---|
| Predicted/Expressed Symptom | NOESY SPECTRA (Cross validation) | |
| Predicted as UAP | 25 | 11 |
| Predicted as DAIP | 4 | 24 |
| Accuracy (%) | 77.6 | |
| Predicted/Expressed Symptom | CPMG SPECTRA (Cross validation) | |
| Predicted as UAP | 28 | 3 |
| Predicted as DAIP | 1 | 32 |
| Accuracy (%) | 93.8 | |
| Predicted/Expressed Symptom | DIFFUSION SPECTRA (Cross validation) | |
| Predicted as UAP | 21 | 12 |
| Predicted as DAIP | 8 | 23 |
| Accuracy (%) | 69.4 | |
| Predicted/Diagnosed | Symptomatic Gallstones | Cholecystitis | Pancreatitis |
|---|---|---|---|
| CPMG SPECTRA (Cross validation) | |||
| Predicted as Symptomatic Gallstones | 9 | 0 | 0 |
| Predicted as Cholecystitis | 0 | 5 | 1 |
| Predicted as Pancreatitis | 0 | 1 | 4 |
| Accuracy (%) | 100 | 88.1 | 90 |
| Predicted/Diagnosed | Intestinal Ischemia | Intestinal Strangulated Obstruction | Intestinal Mechanical Obstruction |
|---|---|---|---|
| CPMG SPECTRA (Cross Validation) | |||
| Predicted as Intestinal ischemia | 6 | 1 | 0 |
| Predicted as Intestinal Strangulated Obstruction | 0 | 5 | 0 |
| Predicted as Intestinal Mechanical Obstruction | 0 | 0 | 11 |
| Accuracy (%) | 95.7 | 90.7 | 100 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takis, P.G.; Taddei, A.; Pini, R.; Grifoni, S.; Tarantini, F.; Bechi, P.; Luchinat, C. Fingerprinting Acute Digestive Diseases by Untargeted NMR Based Metabolomics. Int. J. Mol. Sci. 2018, 19, 3288. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19113288
Takis PG, Taddei A, Pini R, Grifoni S, Tarantini F, Bechi P, Luchinat C. Fingerprinting Acute Digestive Diseases by Untargeted NMR Based Metabolomics. International Journal of Molecular Sciences. 2018; 19(11):3288. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19113288
Chicago/Turabian StyleTakis, Panteleimon G., Antonio Taddei, Riccardo Pini, Stefano Grifoni, Francesca Tarantini, Paolo Bechi, and Claudio Luchinat. 2018. "Fingerprinting Acute Digestive Diseases by Untargeted NMR Based Metabolomics" International Journal of Molecular Sciences 19, no. 11: 3288. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19113288