Structural Studies of the 3′,3′-cGAMP Riboswitch Induced by Cognate and Noncognate Ligands Using Molecular Dynamics Simulation
Abstract
:1. Introduction
2. Results
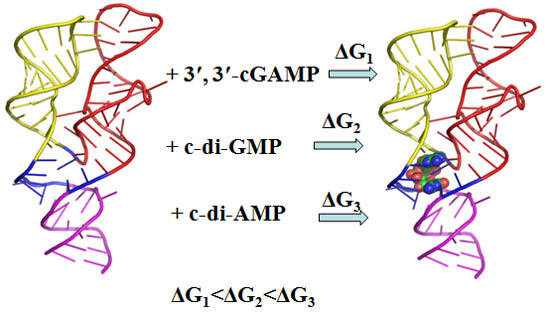
2.1. Energy Calculations Revealed the Preferential Binding of the 3′,3′-cGAMP Riboswitch to 3′,3′-cGAMP over c-di-GMP and c-di-AMP
2.2. Interaction Analysis between the 3′,3′-cGAMP Riboswitch and Ligands
2.3. Structural Characteristics of the 3′,3′-cGAMP Riboswitch in the 3′,3′-cGAMP-Bound and c-di-GMP-Bound State
2.4. Structural Characteristics of the 3′,3′-cGAMP Riboswitch in the Ligand-Free and 3′,3′-cGAMP-Bound State
2.5. Principal Component Analysis of the 3′,3′-cGAMP Riboswitch in the Ligand-Free and 3′,3′-cGAMP-Bound State
2.6. Allosteric Communication of the 3′,3′-cGAMP Riboswitch from Binding Sites Pocket to the P1 Helix
3. Discussion
Allosteric Communication Network from 3′,3′-cGAMP Binding Sites to P1 Helix
4. Materials and Methods
4.1. Initial Structures
4.2. Ligand Force Field Parameter
4.3. Molecular Dynamics Simulation Protocols
4.4. Principal Component Analysis
4.5. Free-Energy Analyses
4.6. Trajectory Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Breaker, R.R. Complex riboswitches. Science 2008, 319, 1795–1797. [Google Scholar] [CrossRef] [PubMed]
- Edwards, T.E.; Klein, D.J.; Ferréd’Amaré, A.R. Riboswitches: Small-molecule recognition by gene regulatory RNAs. Curr. Opin. Struct. Biol. 2007, 17, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Coppins, R.; Hall, K.; Groisman, E. The intricate world of riboswitches. Curr. Opin. Microbiol. 2007, 10, 176–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serganov, A.; Patel, D.J. Ribozymes, riboswitches and beyond: Regulation of gene expression without proteins. Nat. Rev. Genet. 2007, 8, 776–790. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.N.; Breaker, R.R. Purine sensing by riboswitches. Biol. Cell 2008, 100, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serganov, A.; Nudler, E. A decade of riboswitches. Cell 2013, 152, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Nahvi, A.; Barrick, J.E.; Breaker, R.R. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 2004, 32, 143–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winkler, W.; Nahvi, A.; Breaker, R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 2002, 419, 952–956. [Google Scholar] [CrossRef] [PubMed]
- Cromie, M.J.; Shi, Y.; Latifi, T.; Groisman, A.E. An RNA sensor for intracellular Mg. Cell 2006, 125, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Rd, D.C.; Wakeman, C.A.; Sieling, C.L.; Baker, S.C.; Irnov, I.; Winkler, W.C. Structure and mechanism of a metal-sensing regulatory RNA. Cell 2007, 130, 878–892. [Google Scholar]
- Furukawa, K.; Ramesh, A.; Zhou, Z.; Weinberg, Z.; Vallery, T.; Winkler, W.C.; Breaker, R.R. Bacterial riboswitches cooperatively bind Ni2+ or Co2+ ions and control expression of heavy metal transporters. Mol. Cell 2015, 57, 1088–1098. [Google Scholar] [CrossRef] [PubMed]
- Baker, J.L.; Sudarsan, N.; Weinberg, Z.; Roth, A.; Stockbridge, R.B.; Breaker, R.R. Widespread genetic switches and toxicity resistance proteins for fluoride. Science 2012, 335, 233–235. [Google Scholar] [CrossRef] [PubMed]
- Mandal, M.; Breaker, R.R. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004, 11, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Winkler, W.C.; Cohen-Chalamish, S.; Breaker, R.R. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. USA 2003, 99, 15908–15913. [Google Scholar] [CrossRef] [PubMed]
- Ames, T.D.; Breaker, R.R. Bacterial aptamers that selectively bind glutamine. RNA Biol. 2011, 8, 82–89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winkler, W.C.; Nahvi, A.; Sudarsan, N.; Barrick, J.E.; Breaker, R.R. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 2002, 99, 15908–15913. [Google Scholar] [CrossRef] [PubMed]
- Mandal, M.; Lee, M.; Barrick, J.E.; Weinberg, Z.; Emilsson, G.M.; Ruzzo, W.L.; Breaker, R.R. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science 2004, 306, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Winkler, W.C.; Nahvi, A.; Roth, A.; Collins, J.A.; Breaker, R.R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 2004, 428, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Green, N.J.; Grundy, F.J.; Henkin, T.M. The T box mechanism: tRNA as a regulatory molecule. FEBS Lett. 2010, 584, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Serganov, A.; Yuan, Y.R.; Pikovskaya, O.; Polonskaia, A.; Malinina, L.; Phan, A.T.; Hobartner, C.; Micura, R.; Breaker, R.R.; Patel, D.J. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 2004, 11, 1729–1741. [Google Scholar] [CrossRef] [PubMed]
- Mirandaríos, J.; Navarro, M.; Soberón, M. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc. Natl. Acad. Sci. USA 2001, 98, 9736–9741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edwards, T.E.; Ferré-D’Amaré, A.R. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure 2006, 14, 1459–1468. [Google Scholar] [CrossRef] [PubMed]
- Serganov, A.; Polonskaia, A.; Phan, A.T.; Breaker, R.R.; Patel, D.J. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature 2006, 441, 1167–1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thore, S.; Leibundgut, M.; Ban, N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science 2006, 312, 1208–1211. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, R.; Grundy, F.; Henkin, T. The SMK box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat. Struct. Mol. Biol. 2006, 13, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.X.; Lee, E.R.; Morales, D.R.; Lim, J.; Breaker, R.R. Riboswitches that sense S-adenosylhomocysteine and activate genes involved in coenzyme recycling. Mol. Cell 2008, 29, 691–702. [Google Scholar] [CrossRef] [PubMed]
- Nahvi, A.; Sudarsan, N.; Ebert, M.S.; Zou, X.; Brown, K.L.; Breaker, R.R. Genetic control by a metabolite binding mRNA. Chem. Biol. 2002, 9, 1043–1049. [Google Scholar] [CrossRef]
- Sudarsan, N.; Lee, E.R.; Weinberg, Z.; Moy, R.H.; Kim, J.N.; Link, K.H.; Breaker, R.R. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 2008, 321, 411–413. [Google Scholar] [CrossRef] [PubMed]
- Conner, J.G.; Zamoranosánchez, D.; Park, J.H.; Sondermann, H.; Yildiz, F.H. The ins and outs of cyclic di-GMP signaling in Vibrio cholerae. Curr. Opin. Microbiol. 2017, 36, 20–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blount, K.F.; Breaker, R.R. Riboswitches as antibacterial drug targets. Nat. Biotechnol. 2006, 24, 1558–1564. [Google Scholar] [CrossRef] [PubMed]
- Nudler, E.; Mironov, A.S. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 2004, 29, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.R.; Baker, J.L.; Weinberg, Z.; Sudarsan, N.; Breaker, R.R. An allosteric self-splicing ribozyme triggered by a bacterial second messenger. Science 2010, 329, 845–848. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.W.; Sudarsan, N.; Furukawa, K.; Weinberg, Z.; Wang, J.X.; Breaker, R.R. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat. Chem. Biol. 2013, 9, 834–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, K.D.; Lipchock, S.V.; Ames, T.D.; Wang, J.; Breaker, R.R.; Strobel, S.A. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat. Struct. Mol. Biol. 2009, 16, 1218–1223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulshina, N.; Baird, N.J.; Ferré-D’Amaré, A.R. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat. Struct. Mol. Biol. 2009, 16, 1212–1218. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.D.; Shanahan, C.A.; Moore, E.L.; Simon, A.C.; Strobel, S.A. Structural basis of differential ligand recognition by two classes of bis-(3′-5′)-cyclic dimeric guanosine monophosphate-binding riboswitches. Proc. Natl. Acad. Sci. USA 2011, 108, 7757–7762. [Google Scholar] [CrossRef] [PubMed]
- Gao, A.; Serganov, A. Structural insights into recognition of c-di-AMP by the ydaO riboswitch. Nat. Chem. Biol. 2014, 10, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Ren, A.; Patel, D.J. c-di-AMP binds the ydaO riboswitch in two pseudo-symmetry-related pockets. Nat. Chem. Biol. 2014, 10, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Ren, A.; Wang, X.C.; Kellenberger, C.A.; Rajashankar, K.R.; Jones, R.A.; Hammond, M.C.; Patel, D.J. Structural basis for molecular discrimination by a 3′,3′-cGAMP sensing riboswitch. Cell Rep. 2015, 11, 1–12. [Google Scholar] [CrossRef]
- Smith, K.D.; Lipchock, S.V.; Livingston, A.L.; Shanahan, C.A.; Strobel, S.A. Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch. Biochemistry 2010, 49, 7351–7359. [Google Scholar] [CrossRef] [PubMed]
- Dror, R.O.; Dirks, R.M.; Grossman, J.P.; Xu, H.; Shaw, D.E. Biomolecular simulation: A computational microscope for molecular biology. Annu. Rev. Biophys. 2012, 41, 429–452. [Google Scholar] [CrossRef] [PubMed]
- Bermudez, M.; Mortier, J.; Rakers, C.; Sydow, D.; Wolber, G. More than a look into a crystal ball: Protein structure elucidation guided by molecular dynamics simulations. Drug Discov. Today 2016, 21, 1799–1805. [Google Scholar] [CrossRef] [PubMed]
- Vangaveti, S.; Ranganathan, S.V.; Chen, A.A. Advances in RNA molecular dynamics: A simulator’s guide to RNA force fields. Wiley Interdiscip. Rev. RNA 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Sponer, J.; Bussi, G.; Krepl, M.; Banas, P.; Bottaro, S.; Cunha, R.A.; Gil-Ley, A.; Pinamonti, G.; Poblete, S.; Jurecka, P.; et al. RNA structural dynamics as captured by molecular simulations: A comprehensive overview. Chem. Rev. 2018, 118, 4177–4338. [Google Scholar] [CrossRef] [PubMed]
- Priyakumar, U.D.; Jr, A.D.M. Role of the adenine ligand on the stabilization of the secondary and tertiary Interactions in the adenine riboswitch. J. Mol. Biol. 2010, 396, 1422–1438. [Google Scholar] [CrossRef] [PubMed]
- Nozinovic, S.; Reining, A.; Kim, Y.B.; Noeske, J.; Kai, S.; Wöhnert, J.; Schwalbe, H. The importance of helix P1 stability for structural pre-organization and ligand binding affinity of the adenine riboswitch aptamer domain. RNA Biol. 2014, 11, 655–656. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Kim, J.; Jha, S.; Aboul-ela, F. A mechanism for S-adenosyl methionine assisted formation of a riboswitch conformation: A small molecule with a strong arm. Nucleic Acids Res. 2009, 37, 6528–6539. [Google Scholar] [CrossRef] [PubMed]
- Stoddard, C.D.; Montange, R.K.; Hennelly, S.P.; Rambo, R.P.; Sanbonmatsu, K.Y.; Batey, R.T. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure 2010, 18, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Hayes, R.L.; Noel, J.K.; Mohanty, U.; Whitford, P.C.; Hennelly, S.P.; Onuchic, J.N.; Sanbonmatsu, K.Y. Magnesium fluctuations modulate RNA dynamics in the SAM-I riboswitch. J. Am. Chem. Soc. 2012, 134, 12043–12053. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Kim, J.; Jha, S.; Aboul-Ela, F. The impact of a ligand binding on strand migration in the SAM-I riboswitch. PLoS Comput. Biol. 2013, 9, e1003069. [Google Scholar] [CrossRef] [PubMed]
- Doshi, U.; Kelley, J.M.; Hamelberg, D. Atomic-level insights into metabolite recognition and specificity of the SAM-II riboswitch. RNA 2012, 18, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Yongjun, W.; Zhihong, L. Folding of SAM-II riboswitch explored by replica-exchange molecular dynamics simulation. J. Theor. Biol. 2015, 365, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Kelley, J.M.; Hamelberg, D. Atomistic basis for the on-off signaling mechanism in SAM-II riboswitch. Nucleic Acids Res. 2010, 38, 1392–1400. [Google Scholar] [CrossRef] [PubMed]
- Banáš, P.; Walter, N.G.; Šponer, J.; Otyepka, M. Protonation states of the key active site residues and structural dynamics of the glmS riboswitch as revealed by molecular dynamics. J. Phys. Chem. B 2010, 114, 8701. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y.; Hamelberg, D. Deciphering the role of glucosamine-6-phosphate in the riboswitch action of glmS ribozyme. RNA 2010, 16, 2455–2463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liberman, J.A.; Suddala, K.C.; Aytenfisu, A.; Chan, D.; Belashov, I.A.; Salim, M.; Mathews, D.H.; Spitale, R.C.; Walter, N.G.; Wedekind, J.E. Structural analysis of a class III preQ1 riboswitch reveals an aptamer distant from a ribosome-binding site regulated by fast dynamics. Proc. Natl. Acad. Sci. USA 2015, 112, 3485–3494. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Zhao, Y.; Chen, C.; Yong, D.; Yi, X. Insights into ligand binding to PreQ 1 riboswitch aptamer from molecular dynamics simulations. PLoS ONE 2014, 9, e92247. [Google Scholar]
- Eichhorn, C.D.; Feng, J.; Suddala, K.C.; Walter, N.G.; Iii, B.C.L.; Al-Hashimi, H.M. Unraveling the structural complexity in a single-stranded RNA tail: Implications for efficient ligand binding in the prequeuosine riboswitch. Nucleic Acids Res. 2012, 40, 1345–1355. [Google Scholar] [CrossRef] [PubMed]
- Banáš, P.; Sklenovský, P.; Wedekind, J.E.; Šponer, J.; Otyepka, M. Molecular mechanism of preQ1 riboswitch action: A molecular dynamics study. J. Phys. Chem. B 2012, 116, 12721–12734. [Google Scholar] [CrossRef] [PubMed]
- Petrone, P.M.; Dewhurst, J.; Tommasi, R.; Whitehead, L.; Pomerantz, A.K. Atomic-scale characterization of conformational changes in the preQ 1 riboswitch aptamer upon ligand binding. J. Mol. Graph. Model. 2011, 30, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Aytenfisu, A.H.; Liberman, J.A.; Wedekind, J.E.; Mathews, D.H. Molecular mechanism for preQ1-II riboswitch function revealed by molecular dynamics. RNA 2015, 21, 1898–1907. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Jiang, C.; Zhang, J.; Ye, W.; Luo, R.; Chen, H.F. Dynamics correlation network for allosteric switching of PreQ1 riboswitch. Sci. Rep. 2016, 6, 31005. [Google Scholar] [CrossRef] [PubMed]
- Villa, A.; Hnert, J.W.; Stock, G. Molecular dynamics simulation study of the binding of purine bases to the aptamer domain of the guanine sensing riboswitch. Nucleic Acids Res. 2009, 37, 4774–4786. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sund, J.; Lind, C.; Åqvist, J. Binding site preorganization and ligand discrimination in the purine riboswitch. J. Phys. Chem. B 2015, 119, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhao, X.; Xie, P.; Hu, J.; Bi, H. Molecular dynamics simulation on the allosteric analysis of the c-di-GMP class I riboswitch induced by ligand binding. J. Mol. Recognit. 2018, e2756. [Google Scholar] [CrossRef] [PubMed]
- Sanbonmatsu, K.Y. Dynamics of riboswitches: Molecular simulations. BBA Gene Regul. Mech. 2014, 1839, 1046–1050. [Google Scholar] [CrossRef] [PubMed]
- Hess, B. Determining the shear viscosity of model liquids from molecular dynamics simulations. J. Chem. Phys. 2002, 116, 209–217. [Google Scholar] [CrossRef]
- Flyvbjerg, H.; Petersen, H.G. Error estimates on averages of correlated data. J. Chem. Phys. 1989, 91, 88–103. [Google Scholar] [CrossRef]
- Chuprina, V.; Rullmann, J.; Lamerichs, R.; Van Boom, J.; Boelens, R.; Kaptein, R. Structure of the complex of lac repressor headpiece and an 11 base-pair half-operator determined by nuclear magnetic resonance spectroscopy and restrained molecular dynamics. J. Mol. Biol. 1993, 234, 446–462. [Google Scholar] [CrossRef] [PubMed]
- Brocklehurst, S.M.; Perham, R.N. Prediction of the three-dimensional structures of the biotinylated domain from yeast pyruvate carboxylase and of the lipoylated H-protein from the pea leaf glycine cleavage system: A new automated method for the prediction of protein tertiary structure. Protein Sci. 1993, 2, 626–639. [Google Scholar] [CrossRef] [PubMed]
- Burkard, M.E.; Kierzek, R.; Turner, D.H. Thermodynamics of unpaired terminal nucleotides on short RNA helixes correlates with stacking at helix termini in larger RNAs. J. Mol. Biol. 1999, 290, 967–982. [Google Scholar] [CrossRef] [PubMed]
- Bayly, C.I.; Cieplak, P.; Cornell, W.; Kollman, P.A. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: The RESP model. J. Phys. Chem. 1993, 97, 10269–10280. [Google Scholar] [CrossRef]
- Frisch, M.; Trucks, G.; Schlegel, H.; Scuseria, G.; Robb, M.; Cheeseman, J.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.; et al. DJ Fox Gaussian 09, Revision D. 01; Gaussian, Inc.: Wallingford, CT, USA, 2009.
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Case, D.A.; Cerutti, D.; Cheatham, T.E., III; Darden, T.A.; Duke, R.E.; Giese, T.J.; Gohlke, H.; Goetz, A.W.; Izadi, S.; Janowski, P.; et al. AMBER 2016; University of California: San Francisco, CA, USA, 2016. [Google Scholar]
- Lee, M.C.; Duan, Y. Distinguish protein decoys by using a scoring function based on a new AMBER force field, short molecular dynamics simulations, and the generalized born solvent model. Proteins 2004, 55, 620–634. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Wu, C.; Chowdhury, S.; Lee, M.C.; Xiong, G.; Zhang, W.; Yang, R.; Cieplak, P.; Luo, R.; Lee, T. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003, 24, 1999–2012. [Google Scholar] [CrossRef] [PubMed]
- Perez, A.; Marchan, I.; Svozil, D.; Sponer, J.; Cheatham, T.E., 3rd; Laughton, C.A.; Orozco, M. Refinement of the AMBER force field for nucleic acids: Improving the description of alpha/gamma conformers. Biophys. J. 2007, 92, 3817–3829. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, Y.; Chen, G. Interaction investigations of HipA binding to HipB dimer and HipB dimer + DNA complex: A molecular dynamics simulation study. J. Mol. Recognit. 2013, 26, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ma, N.; Wang, Y.; Chen, G. Molecular dynamics simulation studies on the positive cooperativity of the Kemptide substrate with protein kinase A induced by the ATP ligand. J. Phys. Chem. B 2014, 118, 1273–1287. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhu, Y.; Wang, Y.; Chen, G. Molecular dynamics simulation on the conformational transition of the Mad2 protein from the open to the closed state. Int. J. Mol. Sci. 2014, 15, 5553–5569. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, J. Molecular dynamics and continuum solvent studies of the stability of PolyG-PolyC and PolyA-PolyT DNA duplexes in solution. J. Biomol. Struct. Dyn. 1998, 16, 265–280. [Google Scholar]
- Kollman, P.A.; Massova, I.; Reyes, C.; Kuhn, B.; Huo, S.; Chong, L.; Lee, M.; Lee, T.; Duan, Y.; Wang, W. Calculating structures and free energies of complex molecules: Combining molecular mechanics and continuum models. Accounts Chem. Res. 2000, 33, 889–897. [Google Scholar] [CrossRef]
- Jayaram, B.; Sprous, D.; Young, M.A.; Beveridge, D.L. Free energy analysis of the conformational preferences of A and B Forms of DNA in solution. J. Am. Chem. Soc. 1998, 120, 10629–10633. [Google Scholar] [CrossRef]
- Srinivasan, J.; Cheatham, T.E., III; Cieplak, P.; Kollman, P.A.; Case, D.A. Continuum solvent studies of the stability of DNA, RNA, and phosphoramidate−DNA helices. J. Am. Chem. Soc. 1998, 120, 9401–9409. [Google Scholar] [CrossRef]
- Sadiq, S.K.; De Fabritiis, G. Explicit solvent dynamics and energetics of HIV-1 protease flap opening and closing. Proteins 2010, 78, 2873–2885. [Google Scholar] [CrossRef] [PubMed]












| Energies | Riboswitch + 3′,3′-cGAMP | Riboswitch + c-di-GMP | Riboswitch + c-di-AMP |
|---|---|---|---|
| Receptor | 3′,3′-cGAMP riboswitch | 3′,3′-cGAMP riboswitch | 3′,3′-cGAMP riboswitch |
| Ligand | 3′,3′-cGAMP | c-di-GMP | c-di-AMP |
| ΔEele | 1349.93 | 2699.95 | 2647.28 |
| ΔEvdw | −81.92 | −78.61 | −67.59 |
| ΔGnp/solv | −7.26 | −7.08 | −6.73 |
| ΔGpb/solv | −1296.48 | −2645.96 | −2587.86 |
| ΔGnp | −89.18 | −85.69 | −74.32 |
| ΔGpb | 53.45 | 54.00 | 59.42 |
| ΔH | −35.73 | −31.69 | −14.90 |
| TΔS | −24.11 | −22.95 | −28.21 |
| ΔGbinding | −11.62 (0.12) a | −8.74 (0.11) a | 13.31 (0.12) a |
| ΔGexp | −9.82 | −8.28 | --- |
| kd | 0.07 uM | 0.93 uM |
| Riboswitch + 3′,3′-cGAMP | Riboswitch + c-di-GMP | ||||
|---|---|---|---|---|---|
| 3′,3′-cGAMP Riboswitch∙∙∙3′,3′-cGAMP | Occupancies (%) | Error | 3′,3′-cGAMP Riboswitch∙∙∙3′,3′-cGAMP | Occupancies (%) | Error |
| (G8)O2′-H∙∙∙O9(3′,3′-cGAMP) | 55.00 | 3.5 | --- | --- | --- |
| (G8)O13-H∙∙∙O10(3′,3′-cGAMP) | 48.44 | 2.1 | --- | --- | --- |
| (A9)O2′∙∙∙H-O12(3′,3′-cGAMP) | 77.12 | 5.0 | --- | --- | --- |
| (A11)N6-H∙∙∙O12(3′,3′-cGAMP) | 91.54 | 1.0 | (A11)N6-H∙∙∙O13(c-di-GMP) | 36.56 | 2.8 |
| (A11)N7∙∙∙H-O12(3′,3′-cGAMP) | 5.12 | 1.4 | --- | --- | --- |
| (A12)N1∙∙∙H-N10(3′,3′-cGAMP) | 99.76 | 0.1 | (A12)N1∙∙∙H-N9(c-di-GMP) | 88.68 | 3.8 |
| (A12)N6-H∙∙∙O3(3′,3′-cGAMP) | 96.50 | 1.4 | (A12)N6-H∙∙∙N10(c-di-GMP) | 78.96 | 3.5 |
| (A12)N6-H ∙∙N7(3′,3′-cGAMP) | 43.88 | 1.3 | (A12) N6-H∙∙∙N9(c-di-GMP) | 30.20 | 4.7 |
| (A12)N6-H∙∙∙P1(3′,3′-cGAMP) | 5.22 | 0.4 | --- | --- | --- |
| (A14)N2-H∙∙∙N3(3′,3′-cGAMP) | 40.22 | 1.2 | (A14)N6-H∙∙∙O1(c-di-GMP) | 80.28 | 3.0 |
| (A14)N1∙∙∙H-N2(3′,3′-cGAMP) | 38.36 | 1.3 | (A14) N6-H∙∙∙N4(c-di-GMP) | 6.48 | 0.8 |
| (A14)N6∙∙∙H-N2(3′,3′-cGAMP) | 6.80 | 2.6 | --- | --- | --- |
| (A14)N6∙∙∙H-N2(3′,3′-cGAMP) | 6.56 | 2.8 | --- | --- | --- |
| (C15)N4-H∙∙∙N3(3′,3′-cGAMP) | 16.70 | 1.4 | --- | --- | --- |
| (C15)N3∙∙∙H-N2(3′,3′-cGAMP) | 6.52 | 2.6 | --- | --- | --- |
| (G40)O2P∙∙∙H-O13(3′,3′-cGAMP) | 27.46 | 1.9 | (G40)O2′∙∙∙H-N2(c-di-GMP) | 58.92 | 2.1 |
| (G40)P∙∙∙H-O13(3′,3′-cGAMP) | 19.46 | 1.7 | --- | --- | --- |
| (A41)N6-H∙∙∙O4(3′,3′-cGAMP) | 99.30 | 0.2 | (A41)N6-H∙∙∙O11(c-di-GMP) | 99.72 | 0.1 |
| (A41)O2P∙∙∙H-O13(3′,3′-cGAMP) | 70.34 | 5.4 | (A41)O2P∙∙∙H-O3(c-di-GMP) | 99.20 | 0.4 |
| (A41)O2′-H∙∙∙N8(3′,3′-cGAMP) | 16.68 | 5.0 | (A41) O2′-H∙∙∙N7(c-di-GMP) | 49.96 | 6.1 |
| --- | --- | --- | (A41)O5′∙∙∙H-N2(c-di-GMP) | 5.72 | 3.1 |
| (A42)N3∙∙∙H-N2(3′,3′-cGAMP) | 22.20 | 1.3 | (A42)O2′∙∙∙H-N2(c-di-GMP) | 21.00 | 3.6 |
| (C75)N3∙∙∙H-N10(3′,3′-cGAMP) | 99.52 | 0.4 | (C75)N4-H∙∙∙N8(c-di-GMP) | 98.84 | 0.9 |
| (C75)N4-H∙∙∙N9(3′,3′-cGAMP) | 98.40 | 0.6 | (C75)N3∙∙∙H-N9(c-di-GMP) | 97.52 | 0.8 |
| (C75)O2∙∙∙H-N10(3′,3′-cGAMP) | 52.02 | 1.8 | (C75)N4-H∙∙∙O14(c-di-GMP) | 60.40 | 7.6 |
| (C75)N4-H∙∙∙O11(3′,3′-cGAMP) | 31.26 | 4.5 | (C75)O2∙∙∙H-N9(c-di-GMP) | 37.24 | 7.0 |
| Systems | PC1 | PC2 | PC3 | PCs a |
|---|---|---|---|---|
| apo riboswitch | 69.85 | 12.34 | 9.01 | 91.20 |
| Riboswitch +3′,3′-cGAMP | 72.07 | 11.50 | 7.39 | 90.96 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Zhao, X.; Zhu, X.; Xie, P.; Chen, G. Structural Studies of the 3′,3′-cGAMP Riboswitch Induced by Cognate and Noncognate Ligands Using Molecular Dynamics Simulation. Int. J. Mol. Sci. 2018, 19, 3527. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19113527
Li C, Zhao X, Zhu X, Xie P, Chen G. Structural Studies of the 3′,3′-cGAMP Riboswitch Induced by Cognate and Noncognate Ligands Using Molecular Dynamics Simulation. International Journal of Molecular Sciences. 2018; 19(11):3527. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19113527
Chicago/Turabian StyleLi, Chaoqun, Xiaojia Zhao, Xiaomin Zhu, Pengtao Xie, and Guangju Chen. 2018. "Structural Studies of the 3′,3′-cGAMP Riboswitch Induced by Cognate and Noncognate Ligands Using Molecular Dynamics Simulation" International Journal of Molecular Sciences 19, no. 11: 3527. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19113527