α-Chymotrypsin Immobilized on a Low-Density Polyethylene Surface Successfully Weakens Escherichia coli Biofilm Formation
Abstract
:1. Introduction
2. Results
2.1. Immobilized α-CT Retains Its Activity
2.2. LDPE-α-CT Reduces Biofilm Biomass
2.3. LDPE-α-CT Reduces Biofilm Biomass without Affecting Cell Viability
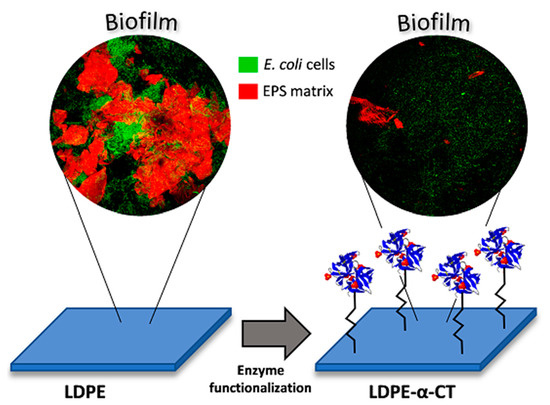
2.4. LDPE-α-CT Affect Biofilm Morphology
3. Discussion
4. Materials and Methods
4.1. Polymeric Surface Preparation
4.2. Evaluation of Immobilized α-CT Activity
4.3. E. coli Strain and Growth Condition
4.4. Biofilm Growth in the CDC Reactor
4.5. Plate Count Viability Assay
4.6. Epifluorescence Microscopy Analysis
4.7. Confocal Laser Scanning Microscopy (CLSM) Analysis
4.8. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Swan, J.S.; Deasy, E.C.; Boyle, M.A.; Russell, R.J.; O’Donnell, M.J.; Coleman, D.C. Elimination of biofilm and microbial contamination reservoirs in hospital washbasin U-bends by automated cleaning and disinfection with electrochemically activated solutions. J. Hosp. Infect. 2016, 94, 169–174. [Google Scholar] [CrossRef]
- Chen, M.; Yu, Q.; Sun, H. Novel strategies for the prevention and treatment of biofilm related infections. Int. J. Mol. Sci. 2013, 14, 18488–18501. [Google Scholar] [CrossRef] [PubMed]
- Lo, J.; Lange, D.; Chew, B.H. Ureteral stents and foley catheters-associated urinary tract infections: The role of coatings and materials in infection prevention. Antibiotics 2014, 3, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Ahire, J.J.; Hattingh, M.; Neveling, D.P.; Dicks, L.M.T. Copper-containing anti-biofilm nanofiber scaffolds as a wound dressing material. PLoS ONE 2016, 11, e0152755. [Google Scholar] [CrossRef] [PubMed]
- Garuglieri, E.; Catto, C.; Villa, F.; Zanchi, R.; Cappitelli, F. Effects of sublethal concentrations of silver nanoparticles on Escherichia coli and Bacillus subtilis under aerobic and anaerobic conditions. Biointerphases 2016, 11, 04B308. [Google Scholar] [CrossRef] [PubMed]
- Pechook, S.; Sudakov, K.; Polishchuk, I.; Ostrov, I.; Zakin, V.; Pokroy, B.; Shemesh, M. Bioinspired passive anti-biofouling surfaces preventing biofilm formation. J. Mater. Chem. B 2015, 3, 1371–1378. [Google Scholar] [CrossRef]
- Costerton, J.W. Introduction to biofilm. Int. J. Antimicrob. Agents 1999, 11, 217–221; discussion 237–219. [Google Scholar] [CrossRef]
- Ventola, C.L. The antibiotic resistance crisis: Part 1: Causes and threats. Pharm. Ther. 2015, 40, 277–283. [Google Scholar]
- Frieri, M.; Kumar, K.; Boutin, A. Antibiotic resistance. J. Infect. Public Health 2017, 10, 369–378. [Google Scholar] [CrossRef]
- Batoni, G.; Maisetta, G.; Esin, S. Antimicrobial peptides and their interaction with biofilms of medically relevant bacteria. Biochim. Biophys. Acta 2016, 1858, 1044–1060. [Google Scholar] [CrossRef]
- Von Eiff, C.; Jansen, B.; Kohnen, W.; Becker, K. Infections associated with medical devices: Pathogenesis, management and prophylaxis. Drugs 2005, 65, 179–214. [Google Scholar] [CrossRef] [PubMed]
- Gharbi, A.; Humblot, V.; Turpin, F.; Pradier, C.M.; Imbert, C.; Berjeaud, J.M. Elaboration of antibiofilm surfaces functionalized with antifungal-cyclodextrin inclusion complexes. FEMS Immunol. Med. Microbiol. 2012, 65, 257–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef] [PubMed]
- Fleming, D.; Rumbaugh, K.P. Approaches to dispersing medical biofilms. Microorganisms 2017, 5, 15. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, J.B. Biofilm matrix-degrading enzymes. Methods Mol. Biol. 2014, 1147, 203–213. [Google Scholar] [CrossRef] [PubMed]
- Elchinger, P.H.; Delattre, C.; Faure, S.; Roy, O.; Badel, S.; Bernardi, T.; Taillefumier, C.; Michaud, P. Immobilization of proteases on chitosan for the development of films with anti-biofilm properties. Int. J. Biol. Macromol. 2015, 72, 1063–1068. [Google Scholar] [CrossRef]
- Stiefel, P.; Mauerhofer, S.; Schneider, J.; Maniura-Weber, K.; Rosenberg, U.; Ren, Q. Enzymes enhance biofilm removal efficiency of cleaners. Antimicrob. Agents Chemother. 2016, 60, 3647–3652. [Google Scholar] [CrossRef]
- Mitrofanova, O.; Mardanova, A.; Evtugyn, V.; Bogomolnaya, L.; Sharipova, M. Effects of Bacillus Serine Proteases on the Bacterial Biofilms. BioMed Res. Int. 2017, 2017, 8525912. [Google Scholar] [CrossRef]
- Sharma, K.; Singh, A.P. Antibiofilm effect of dnase against single and mixed species biofilm. Foods 2018, 7, 42. [Google Scholar] [CrossRef]
- Homaei, A.A.; Sariri, R.; Vianello, F.; Stevanato, R. Enzyme immobilization: An update. J. Chem. Biol. 2013, 6, 185–205. [Google Scholar] [CrossRef]
- Villa, F.; Cappitelli, F. Plant-derived bioactive compounds at sub-lethal concentrations: Towards smart biocide-free antibiofilm strategies. Phytochem. Rev. 2013, 12, 245–254. [Google Scholar] [CrossRef]
- Kaplan, J.B. Biofilm dispersal: Mechanisms, clinical implications, and potential therapeutic uses. J. Dent. Res. 2010, 89, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Azeredo, J.; Azevedo, N.F.; Briandet, R.; Cerca, N.; Coenye, T.; Costa, A.R.; Desvaux, M.; Di Bonaventura, G.; Hebraud, M.; Jaglic, Z.; et al. Critical review on biofilm methods. Crit. Rev. Microbiol. 2017, 43, 313–351. [Google Scholar] [CrossRef] [PubMed]
- Spadoni-Andreani, E.S.; Magagnin, L.; Secundo, F. Preparation and comparison of hydrolase-coated plastics. Chemistryselect 2016, 1, 1490–1495. [Google Scholar] [CrossRef]
- Stepankova, V.; Bidmanova, S.; Koudelakova, T.; Prokop, Z.; Chaloupkova, R.; Damborsky, J. Strategies for stabilization of enzymes in organic solvents. ACS Catal. 2013, 3, 2823–2836. [Google Scholar] [CrossRef]
- Sastri, V.R. Commodity thermoplastics: Polyvinyl chloride, polyolefin, and polystyrene. In Plastics in Medical Devices: Properties, Requirements, and Applications, 2nd ed.; Sastri, V.R., Ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2010; pp. 73–119. ISBN 9780323265638. [Google Scholar]
- Jordan, J.L.; Casem, D.T.; Bradley, J.M.; Dwivedi, A.K.; Brown, E.N.; Jordan, C.W. Mechanical properties of low density polyethylene. J. Dyn. Behav. Mater. 2016, 2, 411–420. [Google Scholar] [CrossRef]
- Unger, S.; Landis, A. Assessing the environmental, human health, and economic impacts of reprocessed medical devices in a Phoenix hospital’s supply chain. J. Clean. Prod. 2016, 112, 1995–2003. [Google Scholar] [CrossRef]
- Raj, B.; Sankar, U.K.; Siddaramaiah. Low density polyethylene/starch blend films for food packaging applications. Adv. Polym. Technol. 2004, 23, 32–45. [Google Scholar] [CrossRef]
- Villa, F.; Secundo, F.; Polo, A.; Cappitelli, F. Immobilized hydrolytic enzymes exhibit antibiofilm activity against Escherichia coli at sub-lethal concentrations. Curr. Microbiol. 2015, 71, 106–114. [Google Scholar] [CrossRef]
- Secundo, F.; Barletta, G.L.; Parini, G.; Roda, G. Effects of stabilizing additives on the activity of alpha-chymotrypsin in organic solvent. J. Mol. Catal. B Enzym. 2012, 84, 128–131. [Google Scholar] [CrossRef]
- Novick, S.J.; Dordick, J.S. Protein-containing hydrophobic coatings and films. Biomaterials 2002, 23, 441–448. [Google Scholar] [CrossRef]
- Zanaroli, G.; Negroni, A.; Calisti, C.; Ruzzi, M.; Fava, F. Selection of commercial hydrolytic enzymes with potential antifouling activity in marine environments. Enzyme Microb. Technol. 2011, 49, 574–579. [Google Scholar] [CrossRef] [PubMed]
- Artini, M.; Papa, R.; Scoarughi, G.L.; Galano, E.; Barbato, G.; Pucci, P.; Selan, L. Comparison of the action of different proteases on virulence properties related to the staphylococcal surface. J. Appl. Microbiol. 2013, 114, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Harris, L.G.; Nigam, Y.; Sawyer, J.; Mack, D.; Pritchard, D.I. Lucilia sericata chymotrypsin disrupts protein adhesin-mediated staphylococcal biofilm formation. Appl. Environ. Microbiol. 2013, 79, 1393–1395. [Google Scholar] [CrossRef] [PubMed]
- Leccese Terraf, M.C.; Tomas, M.S.J.; Rault, L.; Le Loir, Y.; Even, S.; Nader-Macias, M.E.F. Biofilms of vaginal Lactobacillus reuteri CRL 1324 and Lactobacillus rhamnosus CRL 1332: Kinetics of formation and matrix characterization. Arch. Microbiol. 2016, 198, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Spadoni-Andreani, E.S.; Villa, F.; Cappitelli, F.; Krasowska, A.; Biniarz, P.; Lukaszewicz, M.; Secundo, F. Coating polypropylene surfaces with protease weakens the adhesion and increases the dispersion of Candida albicans cells. Biotechnol. Lett. 2017, 39, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Mohamad, N.R.; Marzuki, N.H.; Buang, N.A.; Huyop, F.; Wahab, R.A. An overview of technologies for immobilization of enzymes and surface analysis techniques for immobilized enzymes. Biotechnol. Equip. 2015, 29, 205–220. [Google Scholar] [CrossRef] [Green Version]
- Saldarriaga Fernández, I.C.S.; van der Mei, H.C.; Lochhead, M.J.; Grainger, D.W.; Busscher, H.J. The inhibition of the adhesion of clinically isolated bacterial strains on multi-component cross-linked poly(ethylene glycol)-based polymer coatings. Biomaterials 2007, 28, 4105–4112. [Google Scholar] [CrossRef]
- Dell’orto, S.; Cattò, C.; Villa, F.; Forlani, F.; Vassallo, E.; Morra, M.; Cappitelli, F.; Villa, S.; Gelain, A. Low density polyethylene functionalized with antibiofilm compounds inhibits Escherichia coli cell adhesion. J. Biomed. Mater. Res. A 2017, 105, 3251–3261. [Google Scholar] [CrossRef]
- Rather, I.A.; Koh, W.Y.; Paek, W.K.; Lim, J. The sources of chemical contaminants in food and their health implications. Front. Pharmacol. 2017, 8, 830. [Google Scholar] [CrossRef]
- Garcia-Galan, C.; Berenguer-Murcia, A.; Fernandez-Lafuente, R.; Rodrigues, R.C. Potential of different enzyme immobilization strategies to improve enzyme performance. Adv. Synth. Catal. 2011, 353, 2885–2904. [Google Scholar] [CrossRef]
- Chae, H.J.; In, M.J.; Kim, E.Y. Optimization of protease immobilization by covalent binding using glutaraldehyde. Appl. Biochem. Biotechnol. 1998, 73, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Gallego, F.; Guisan, J.M.; Betancor, L. Glutaraldehyde-mediated protein immobilization. Methods Mol. Biol. 2013, 1051, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, A.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Glutaraldehyde in bio-catalysts design: A useful crosslinker and a versatile tool in enzyme immobilization. RSC Adv. 2014, 4, 1583–1600. [Google Scholar] [CrossRef]
- Li, D.F.; Ding, H.C.; Zhou, T. Covalent immobilization of mixed proteases, trypsin and chymotrypsin, onto modified polyvinyl chloride microspheres. J. Agric. Food Chem. 2013, 61, 10447–10453. [Google Scholar] [CrossRef] [PubMed]
- Xiao, P.; Lv, X.F.; Deng, Y.L. Immobilization of chymotrypsin on silica beads based on high affinity and specificity aptamer and its applications. Anal. Lett. 2012, 45, 1264–1273. [Google Scholar] [CrossRef]
- Onyshchenko, I.; De Geyter, N.; Nikiforov, A.Y.; Morent, R. Atmospheric pressure plasma penetration inside flexible polymeric tubes. Plasma Process. Polym. 2015, 12, 271–284. [Google Scholar] [CrossRef]
- Pedroni, M.; Morandi, S.; Silvetti, T.; Cremona, A.; Gittini, G.; Nardone, A.; Pallotta, F.; Brasca, M.; Vassallo, E. Bacteria inactivation by atmospheric pressure plasma jet treatment. J. Vac. Sci. Technol. B 2018, 36, 01A107. [Google Scholar] [CrossRef]
- Lin, W.S.; Niu, B.; Yi, J.L.; Deng, Z.R.; Song, J.; Chen, Q. Toxicity and metal corrosion of glutaraldehyde-didecyldimethylammonium bromide as a disinfectant agent. Biomed Res. Int. 2018, 2018, 9814209. [Google Scholar] [CrossRef]
- Gilmore, B.F.; Hamill, T.M.; Jones, D.S.; Gorman, S.P. Validation of the CDC Biofilm reactor as a dynamic model for assessment of encrustation formation on urological device materials. J. Biomed. Mater. Res. B 2010, 93B, 128–140. [Google Scholar] [CrossRef]
- Williams, D.L.; Woodbury, K.L.; Haymond, B.S.; Parker, A.E.; Bloebaum, R.D. A Modified CDC biofilm reactor to produce mature biofilms on the surface of peek membranes for an in vivo animal model application. Curr. Microbiol. 2011, 62, 1657–1663. [Google Scholar] [CrossRef] [PubMed]
- Heydorn, A.; Nielsen, A.T.; Hentzer, M.; Sternberg, C.; Givskov, M.; Ersbøll, B.K.; Molin, S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 2000, 146, 2395–2407. [Google Scholar] [CrossRef] [PubMed]
- Mangwani, N.; Shukla, S.K.; Kumari, S.; Das, S.; Rao, T.S. Effect of biofilm parameters and extracellular polymeric substance composition on polycyclic aromatic hydrocarbon degradation. RSC Adv. 2016, 6, S7540–S7551. [Google Scholar] [CrossRef]
- Bester, E.; Kroukamp, O.; Hausner, M.; Edwards, E.A.; Wolfaardt, G.M. Biofilm form and function: Carbon availability affects biofilm architecture, metabolic activity and planktonic cell yield. J. Appl. Microbiol. 2011, 110, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Danese, P.N.; Pratt, L.A.; Dove, S.L.; Kolter, R. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 2000, 37, 424–432. [Google Scholar] [CrossRef] [PubMed]
- Ulett, G.C.; Valle, J.; Beloin, C.; Sherlock, O.; Ghigo, J.M.; Schembri, M.A. Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect. Immun. 2007, 75, 3233–3244. [Google Scholar] [CrossRef]
- Vo, J.L.; Ortiz, G.C.M.; Subedi, P.; Keerthikumar, S.; Mathivanan, S.; Paxman, J.J.; Heras, B. Autotransporter Adhesins in Escherichia coli Pathogenesis. Proteomics 2017, 17, 1600431. [Google Scholar] [CrossRef]
- Hobley, L.; Harkins, C.; MacPhee, C.E.; Stanley-Wall, N.R. Giving structure to the biofilm matrix: An overview of individual strategies and emerging common themes. FEMS Microbiol. Rev. 2015, 39, 649–669. [Google Scholar] [CrossRef]
- Kikuchi, T.; Mizunoe, Y.; Takade, A.; Naito, S.; Yoshida, S. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol. Immunol. 2005, 49, 875–884. [Google Scholar] [CrossRef]
- Evans, M.L.; Chapman, M.R. Curli biogenesis: Order out of disorder. Biochim. Biophys. Acta 2014, 1843, 1551–1558. [Google Scholar] [CrossRef]
- Hufnagel, D.A.; Depas, W.H.; Chapman, M.R. The biology of the Escherichia coli extracellular matrix. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef] [PubMed]
- Taglialegna, A.; Lasa, I.; Valle, J. Amyloid structures as biofilm matrix scaffolds. J. Bacteriol. 2016, 198, 2579–2588. [Google Scholar] [CrossRef] [PubMed]
- Reichhardt, C.; Cegelski, L. The Congo red derivative FSB binds to curli amyloid fibers and specifically stains curliated E. coli. PLoS ONE 2018, 13, e0203226. [Google Scholar] [CrossRef] [PubMed]
- Li, B.Y.; Huang, Q.; Cui, A.L.; Liu, X.L.; Hou, B.; Zhang, L.Y.; Liu, M.; Meng, X.R.; Li, S.W. Overexpression of outer membrane protein X (OmpX) compensates for the effect of TolC inactivation on biofilm formation and curli production in extraintestinal pathogenic Escherichia coli (ExPEC). Front. Cell. Infect. Microbiol. 2018, 8, 208. [Google Scholar] [CrossRef] [PubMed]
- Jain, N.; Aden, J.; Nagamatsu, K.; Evans, M.L.; Li, X.Y.; McMichael, B.; Ivanova, M.I.; Almqvist, F.; Buxbaum, J.N.; Chapman, M.R. Inhibition of curli assembly and Escherichia coli biofilm formation by the human systemic amyloid precursor transthyretin. Proc. Natl. Acad. Sci. USA 2017, 114, 12184–12189. [Google Scholar] [CrossRef] [PubMed]
- Fong, J.N.C.; Yildiz, F.H. Biofilm matrix proteins. Microbiol. Spectr. 2015, 3, 2. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Leroy, C.; Delbarre-Ladrat, C.; Ghillebaert, F.; Compere, C.; Combes, D. Effects of commercial enzymes on the adhesion of a marine biofilm-forming bacterium. Biofouling 2008, 24, 11–22. [Google Scholar] [CrossRef]
- Lister, J.L.; Horswill, A.R. Staphylococcus aureus biofilms: Recent developments in biofilm dispersal. Front. Cell. Infect. Microbiol. 2014, 4, 178. [Google Scholar] [CrossRef]
- Davey, M.E.; O’Toole, G.A. Microbial biofilms: From ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 2000, 64, 847–867. [Google Scholar] [CrossRef]
- Shukla, S.K.; Rao, T.S. Staphylococcus aureus biofilm removal by targeting biofilm-associated extracellular proteins. Indian J. Med. Res. 2017, 146, 1–8. [Google Scholar] [CrossRef]
- Estrela, A.B.; Abraham, W.R. Combining biofilm-controlling compounds and antibiotics as a promising new way to control biofilm infections. Pharmaceuticals 2010, 3, 1374–1393. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.O.; Vieira, M.J. Effects of the interactions between glutaraldehyde and the polymeric matrix on the efficacy of the biocide against Pseudomonas fluorescens biofilms. Biofouling 2001, 17, 93–101. [Google Scholar] [CrossRef]
- Simoes, L.C.; Lemos, M.; Araujo, P.; Pereira, A.M.; Simoes, M. The effects of glutaraldehyde on the control of single and dual biofilms of Bacillus cereus and Pseudomonas fluorescens. Biofouling 2011, 27, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Sehmi, S.K.; Allan, E.; MacRobert, A.J.; Parkin, I. The bactericidal activity of glutaraldehyde-impregnated polyurethane. Microbiologyopen 2016, 5, 891–897. [Google Scholar] [CrossRef] [Green Version]
- Tran, V.N.; Dasagrandhi, C.; Truong, V.G.; Kim, Y.M.; Kang, H.W. Antibacterial activity of Staphylococcus aureus biofilm under combined exposure of glutaraldehyde, near-infrared light, and 405-nm laser. PLoS ONE 2018, 13, e0202821. [Google Scholar] [CrossRef]
- Migneault, I.; Dartiguenave, C.; Bertrand, M.J.; Waldron, K.C. Glutaraldehyde: Behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. Biotechniques 2004, 37, 790–796. [Google Scholar] [CrossRef]
- Silva, C.; Sousa, F.; Bitz, G.G.; Cavaco-Paulo, A. Chemical modifications on proteins using glutaraldehyde. Food Technol. Biotechnol. 2004, 42, 51–56. [Google Scholar]
- Maillard, J.Y. Bacterial target sites for biocide action. J. Appl. Microbiol. 2002, 92, S16–S27. [Google Scholar] [CrossRef]
- Van Houdt, R.; Michiels, C.W. Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res. Microbiol. 2005, 156, 626–633. [Google Scholar] [CrossRef]
- Bhushan, B.; Pal, A.; Jain, V. Improved enzyme catalytic characteristics upon glutaraldehyde cross-linking of alginate entrapped xylanase isolated from Aspergillus flavus MTCC 9390. Enzyme Res. 2015, 2015, 210784. [Google Scholar] [CrossRef] [PubMed]
- Faucher, S.P.; Charette, S.J. Editorial on: Bacterial pathogens in the non-clinical environment. Front. Microbiol. 2015, 6, 331. [Google Scholar] [CrossRef] [PubMed]
- Cattò, C.; Grazioso, G.; Dell’Orto, S.; Gelain, A.; Villa, S.; Marzano, V.; Vitali, A.; Villa, F.; Cappitelli, F.; Forlani, F. The response of Escherichia coli biofilm to salicylic acid. Biofouling 2017, 33, 235–251. [Google Scholar] [CrossRef] [PubMed]
- Cattò, C.; James, G.; Villa, F.; Villa, S.; Cappitelli, F. Zosteric acid and salicylic acid bound to a low density polyethylene surface successfully control bacterial biofilm formation. Biofouling 2018, 34, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.W.; Fragkopoulos, A.A.; Marquez, S.M.; Kim, H.D.; Angelini, T.E.; Fernandez-Nieves, A. Biofilm formation in geometries with different surface curvature and oxygen availability. New J. Phys. 2015, 17, 033017. [Google Scholar] [CrossRef] [Green Version]
- Talevi, A. Multi-target pharmacology: Possibilities and limitations of the “skeleton key approach” from a medicinal chemist perspective. Front. Pharmacol. 2015, 6, 205. [Google Scholar] [CrossRef]
- Pfalzgraff, A.; Brandenburg, K.; Weindl, G. Antimicrobial peptides and their therapeutic potential for bacterial skin infections and wounds. Front. Pharmacol. 2018, 9, 281. [Google Scholar] [CrossRef]
- Ramsay, R.R.; Popovic-Nikolic, M.R.; Nikolic, K.; Uliassi, E.; Bolognesi, M.L. A perspective on multi-target drug discovery and design for complex diseases. Clin. Transl. Med. 2018, 7. [Google Scholar] [CrossRef] [Green Version]
- Cappitelli, F.; Polo, A.; Villa, F. Biofilm formation in food processing environments is still poorly understood and controlled. Food Eng. Rev. 2014, 6, 29–42. [Google Scholar] [CrossRef]
- Cui, J.H.; Ren, B.; Tong, Y.J.; Dai, H.Q.; Zhang, L.X. Synergistic combinations of antifungals and anti-virulence agents to fight against Candida albicans. Virulence 2015, 6, 362–371. [Google Scholar] [CrossRef]
- Miquel, S.; Lagrafeuille, R.; Souweine, B.; Forestier, C. Anti-biofilm Activity as a Health Issue. Front. Microbiol. 2016, 7, 592. [Google Scholar] [CrossRef] [PubMed]



| Parameter | LDPE | LDPE-GA | LDPE-α-CT |
|---|---|---|---|
| Thickness (µm) | 20.5 ± 5.0 a | 19.1 ± 4.9 a | 3.7 ± 1.1 b (−81.8 ± 16.7) |
| Roughness | 0.25 ± 0.03 a | 0.22 ± 0.02 b (−11.1 ± 1.0) | 0.21 ± 0.04 b (−13.8 ± 2.8) |
| Substratum coverage (%) | 72.3 ± 3.8 a | 62.3 ± 1.2 b (−13.8 ± 1.7) | 26.7 ± 1.3 c (−63.1 ± 1.8) |
| Total bio-volume (µm3 µm−2) | 89.4 ± 20.1 a | 76.3 ± 21.2 a | 19.7 ± 4.1 b (−78.0 ± 16.1) |
| Cells bio-volume (µm3 µm−2) | 23.7 ± 4.0 a | 18.5 ± 0.9 b (−21.8 ± 1.0) | 6.7 ± 0.3 c (−71.7 ± 3.4) |
| Polysaccharide matrix bio-volume (µm3 µm−2) | 65.8 ± 7.8 a | 57.8 ± 14.9 a | 13.0 ± 3.8 b (−80.2 ± 23.2) |
| Matrix/cells bio-volume ratio | 2.8 ± 0.6 a | 3.1 ± 0.8 a | 1.9 ± 0.5 a |
| Surface/bio-volume (µm2 µm−3) × 10−2 | 1.1 ± 0.3 a | 1.3 ± 0.4 a | 8.7 ± 1.7 b (+7.8 ± 1.6-fold) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cattò, C.; Secundo, F.; James, G.; Villa, F.; Cappitelli, F. α-Chymotrypsin Immobilized on a Low-Density Polyethylene Surface Successfully Weakens Escherichia coli Biofilm Formation. Int. J. Mol. Sci. 2018, 19, 4003. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19124003
Cattò C, Secundo F, James G, Villa F, Cappitelli F. α-Chymotrypsin Immobilized on a Low-Density Polyethylene Surface Successfully Weakens Escherichia coli Biofilm Formation. International Journal of Molecular Sciences. 2018; 19(12):4003. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19124003
Chicago/Turabian StyleCattò, Cristina, Francesco Secundo, Garth James, Federica Villa, and Francesca Cappitelli. 2018. "α-Chymotrypsin Immobilized on a Low-Density Polyethylene Surface Successfully Weakens Escherichia coli Biofilm Formation" International Journal of Molecular Sciences 19, no. 12: 4003. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19124003