Glucosinolate Profiling and Expression Analysis of Glucosinolate Biosynthesis Genes Differentiate White Mold Resistant and Susceptible Cabbage Lines
Abstract
:1. Introduction
2. Results
2.1. Resistance of Different Cabbage Lines to S. sclerotiorum
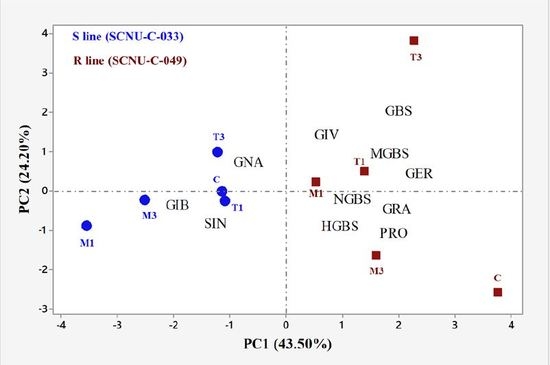
2.2. Overview of Individual GSL Profiles in Cabbage Lines
2.3. Upregulation of MYB28-Bol017019, MYB34-Bol017062, ST5a-Bol026200, ST5a-Bol039395, ST5b-Bol026202 and ST5c-Bol030757 in the R Line after Inoculation
2.4. Upregulation of Transcription Factor-Related Genes and GSL Biosynthesis Genes in the S Line after Inoculation
2.5. Upregulation of Transcription-Factor-Related Genes and GSL Biosynthesis Genes in Both R and S Lines
2.6. Correlation between the Levels of Individual GSLs and the Expression Level of GSL Biosynthesis Pathway Genes Induced by S. sclerotiorum in the R and S Lines
3. Discussion
3.1. Resistance of Cabbage Lines to S. sclerotiorum
3.2. Levels of Total GSLs, GIV, and GBS Were Related to White Mold Resistance
3.3. Increased Expression of ST5b-Bol026202 and ST5c-Bol030757 Led to Increased GIV in the R Line
3.4. Increased Levels of Aliphatic GIV and Indole GBS Were Associated with White Mold Resistance
3.5. Expression of MYB28 and MYB34 Likely Induced Expression of GSL Biosynthesis Genes, Leading to Increases in GBS in the R Line
3.6. Accumulation of Indole GBS in the R Line Was Activated by Increased Expression of GSL Biosynthesis Genes
3.7. Accumulation of Aliphatic GIV with Expression of ST5b-Bol026202 and ST5c-Bol030757 and Indole GBS with ST5a-Bol026200 and ST5a-Bol039395 May Play a Role in Resistance
3.8. Association of GSL Biosynthesis Genes and Accumulation of Individual GSLs in the S Line
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Inoculum Preparation
4.3. Inoculation Technique and Disease Assessment
4.4. Leaf Sampling and Preparation for HPLC and Gene Expression Analysis
4.5. GSL Content Measurements
4.6. Primer Design for Expression Analysis of GSL Biosynthesis Genes
4.7. cDNA Synthesis and RT-qPCR Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Boland, G.; Hall, R. Index of plant hosts of Sclerotinia sclerotiorum. Can. J. Plant Pathol. 1994, 16, 93–108. [Google Scholar] [CrossRef]
- Purdy, L.H. Sclerotinia sclerotiorum: History, diseases and symptomatology, host range, geographic distribution, and impact. Phytopathology 1979, 69, 875–880. [Google Scholar] [CrossRef]
- Jamaux, I.; Gelie, B.; Lamarque, C. Early stages of infection of rapeseed petals and leaves by Sclerotinia sclerotiorum revealed by scanning electron microscopy. Plant Pathol. 1995, 44, 22–30. [Google Scholar] [CrossRef]
- Del Río, L.; Bradley, C.; Henson, R.; Endres, G.; Hanson, B.; McKay, K.; Halvorson, M.; Porter, P.; Le Gare, D.; Lamey, H. Impact of sclerotinia stem rot on yield of canola. Plant Dis. 2007, 91, 191–194. [Google Scholar] [CrossRef]
- Zhao, J.; Peltier, A.; Meng, J.; Osborn, T.; Grau, C. Evaluation of sclerotinia stem rot resistance in oilseed Brassica napus using a petiole inoculation technique under greenhouse conditions. Plant Dis. 2004, 88, 1033–1039. [Google Scholar] [CrossRef]
- Wu, J.; Cai, G.; Tu, J.; Li, L.; Liu, S.; Luo, X.; Zhou, L.; Fan, C.; Zhou, Y. Identification of qtls for resistance to sclerotinia stem rot and bnac. Igmt5. A as a candidate gene of the major resistant QTL SRC6 in Brassica napus. PLoS ONE 2013, 8, e67740. [Google Scholar]
- Garg, H.; Atri, C.; Sandhu, P.S.; Kaur, B.; Renton, M.; Banga, S.K.; Singh, H.; Singh, C.; Barbetti, M.J.; Banga, S.S. High level of resistance to Sclerotinia sclerotiorum in introgression lines derived from hybridization between wild crucifers and the crop Brassica species B. napus and B. juncea. Field Crops Res. 2010, 117, 51–58. [Google Scholar] [CrossRef]
- Lefol, C.; Seguin-Swartz, G.; Morrall, R. Resistance to Sclerotinia sclerotiorum in a weed related to canola. Can. J. Plant Pathol. 1997, 19, 113. [Google Scholar]
- Chen, H.-F.; Wang, H.; Li, Z.-Y. Production and genetic analysis of partial hybrids in intertribal crosses between Brassica species (B. rapa, B. napus) and capsella bursa-pastoris. Plant Cell Rep. 2007, 26, 1791–1800. [Google Scholar] [CrossRef]
- Mei, J.; Qian, L.; Disi, J.; Yang, X.; Li, Q.; Li, J.; Frauen, M.; Cai, D.; Qian, W. Identification of resistant sources against Sclerotinia sclerotiorum in Brassica species with emphasis on B. oleracea. Euphytica 2011, 177, 393–399. [Google Scholar] [CrossRef]
- Bennett, R.N.; Wallsgrove, R.M. Secondary metabolites in plant defense mechanisms. New Phytol. 1994, 127, 617–633. [Google Scholar] [CrossRef]
- Kliebenstein, D. Secondary metabolites and plant/environment interactions: A view through Arabidopsis thaliana tinged glasses. Plant Cell Environ. 2004, 27, 675–684. [Google Scholar] [CrossRef]
- Lattanzio, V.; Lattanzio, V.M.; Cardinali, A. Role of phenolics in the resistance mechanisms of plants against fungal pathogens and insects. Phytochem. Adv. Res. 2006, 661, 23–67. [Google Scholar]
- Wink, M. Plant breeding: Importance of plant secondary metabolites for protection against pathogens and herbivores. Theor. Appl. Genet. 1988, 75, 225–233. [Google Scholar] [CrossRef]
- Fahey, J.W.; Zalcmann, A.T.; Talalay, P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 2001, 56, 5–51. [Google Scholar] [CrossRef]
- Mithen, R. Glucosinolates-biochemistry, genetics and biological activity. Plant Growth Regul. 2001, 34, 91–103. [Google Scholar] [CrossRef]
- Bekaert, M.; Edger, P.P.; Hudson, C.M.; Pires, J.C.; Conant, G.C. Metabolic and evolutionary costs of herbivory defense: Systems biology of glucosinolate synthesis. New Phytol. 2012, 196, 596–605. [Google Scholar] [CrossRef]
- Aires, A.; Mota, V.; Saavedra, M.; Rosa, E.; Bennett, R. The antimicrobial effects of glucosinolates and their respective enzymatic hydrolysis products on bacteria isolated from the human intestinal tract. J. Appl. Microbiol. 2009, 106, 2086–2095. [Google Scholar] [CrossRef] [Green Version]
- Giamoustaris, A.; Mithen, R. The effect of modifying the glucosinolate content of leaves of oilseed rape (Brassica napus ssp. oleifera) on its interaction with specialist and generalist pests. Ann. Appl. Biol. 1995, 126, 347–363. [Google Scholar]
- Manici, L.M.; Lazzeri, L.; Palmieri, S. In vitro fungitoxic activity of some glucosinolates and their enzyme-derived products toward plant pathogenic fungi. J. Agric. Food Chem. 1997, 45, 2768–2773. [Google Scholar] [CrossRef]
- Agerbirk, N.; Olsen, C.E.; Sorensen, H. Initial and final products, nitriles, and ascorbigens produced in myrosinase-catalyzed hydrolysis of indole glucosinolates. J. Agric. Food Chem. 1998, 46, 1563–1571. [Google Scholar] [CrossRef]
- Brader, G.; Tas, E.; Palva, E.T. Jasmonate-dependent induction of indole glucosinolates in Arabidopsis by culture filtrates of the nonspecific pathogen Erwinia carotovora. Plant Physiol. 2001, 126, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Tierens, K.F.-J.; Thomma, B.P.; Brouwer, M.; Schmidt, J.; Kistner, K.; Porzel, A.; Mauch-Mani, B.; Cammue, B.P.; Broekaert, W.F. Study of the role of antimicrobial glucosinolate-derived isothiocyanates in resistance of Arabidopsis to microbial pathogens. Plant Physiol. 2001, 125, 1688–1699. [Google Scholar] [CrossRef] [PubMed]
- Barth, C.; Jander, G. Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J. 2006, 46, 549–562. [Google Scholar] [CrossRef] [PubMed]
- Stotz, H.U.; Sawada, Y.; Shimada, Y.; Hirai, M.Y.; Sasaki, E.; Krischke, M.; Brown, P.D.; Saito, K.; Kamiya, Y. Role of camalexin, indole glucosinolates, and side chain modification of glucosinolate-derived isothiocyanates in defense of Arabidopsis against Sclerotinia sclerotiorum. Plant J. 2011, 67, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Calmes, B.; N’Guyen, G.; Dumur, J.; Brisach, C.A.; Campion, C.; Iacomi, B.; Pigné, S.; Dias, E.; Macherel, D.; Guillemette, T. Glucosinolate-derived isothiocyanates impact mitochondrial function in fungal cells and elicit an oxidative stress response necessary for growth recovery. Front. Plant Sci. 2015, 6, 414. [Google Scholar] [CrossRef] [PubMed]
- Hogge, L.; Reed, D.; Underhill, E.; Haughn, G. Hplc separation of glucosinolates from leaves and seeds of Arabidopsis thaliana and their identification using thermospray liquid chramatography/mass spectrometry. J. Chromatogr. Sci. 1988, 26, 551–556. [Google Scholar] [CrossRef]
- Mithen, R.; Raybould, A.; Giamoustaris, A. Divergent selection for secondary metabolites between wild populations of Brassica oleracea and its implications for plant-herbivore interactions. Heredity 1995, 75, 472. [Google Scholar] [CrossRef]
- Benderoth, M.; Textor, S.; Windsor, A.J.; Mitchell-Olds, T.; Gershenzon, J.; Kroymann, J. Positive selection driving diversification in plant secondary metabolism. Proc. Natl. Acad. Sci. USA 2006, 103, 9118–9123. [Google Scholar] [CrossRef] [Green Version]
- Hopkins, R.J.; van Dam, N.M.; van Loon, J.J. Role of glucosinolates in insect-plant relationships and multitrophic interactions. Annu. Rev. Entomol. 2009, 54, 57–83. [Google Scholar] [CrossRef]
- Schnug, E.; Haneklaus, S.; Murphy, D. Impact of sulphur fertilization on fertilizer nitrogen efficiency. Sulphur Agricult. 1993, 17, 12. [Google Scholar]
- Krumbein, A.; Schonhof, I.; Rühlmann, J.; Widell, S. Influence of sulphur and nitrogen supply on flavour and health-affecting compounds in Brassicaceae. In Plant Nutrition; Springer: Dordrecht, The Netherlands, 2001; pp. 294–295. [Google Scholar]
- Salac, I.; Haneklaus, S.; Bloem, E.; Booth, E.; Sutherland, K.; Walker, K.; Schnug, E. Influence of sulfur fertilization on sulfur metabolites, disease incidence and severity of fungal pathogens in oilseed rape in Scotland. Landbauforschung Vôlkenrode 2006, 56, 1–4. [Google Scholar]
- Schonhof, I.; Blankenburg, D.; Müller, S.; Krumbein, A. Sulfur and nitrogen supply influence growth, product appearance, and glucosinolate concentration of broccoli. J. Plant Nutr. Soil Sci. 2007, 170, 65–72. [Google Scholar] [CrossRef]
- Robin, A.H.K.; Yi, G.-E.; Laila, R.; Yang, K.; Park, J.-I.; Kim, H.R.; Nou, I.-S. Expression profiling of glucosinolate biosynthetic genes in Brassica oleracea L. var. capitata inbred lines reveals their association with glucosinolate content. Molecules 2016, 21, 787. [Google Scholar]
- Robin, A.H.K.; Yi, G.-E.; Laila, R.; Hossain, M.R.; Park, J.-I.; Kim, H.R.; Nou, I.-S. Leptosphaeria maculans alters glucosinolate profiles in blackleg disease-resistant and-susceptible cabbage lines. Front. Plant Sci. 2017, 8, 1769. [Google Scholar] [CrossRef] [PubMed]
- Robin, A.H.K.; Hossain, M.R.; Park, J.-I.; Kim, H.R.; Nou, I.-S. Glucosinolate profiles in cabbage genotypes influence the preferential feeding of diamondback moth (plutella xylostella). Front. Plant Sci. 2017, 8, 1244. [Google Scholar] [CrossRef] [PubMed]
- Abuyusuf, M.; Robin, A.; Kim, H.-T.; Islam, M.; Park, J.-I.; Nou, I.-S. Altered glucosinolate profiles and expression of glucosinolate biosynthesis genes in ringspot-resistant and susceptible cabbage lines. Int. J. Mol. Sci. 2018, 19, 2833. [Google Scholar] [CrossRef] [PubMed]
- Rask, L.; Andréasson, E.; Ekbom, B.; Eriksson, S.; Pontoppidan, B.; Meijer, J. Myrosinase: Gene family evolution and herbivore defense in Brassicaceae. Plant Mol. Biol. 2000, 42, 93–114. [Google Scholar] [CrossRef]
- Wittstock, U.; Halkier, B.A. Glucosinolate research in the Arabidopsis era. Trends Plant Sci. 2002, 7, 263–270. [Google Scholar] [CrossRef]
- Agrawal, A.A.; Kurashige, N.S. A role for isothiocyanates in plant resistance against the specialist herbivore pieris rapae. J. Chem. Ecol. 2003, 29, 1403–1415. [Google Scholar] [CrossRef]
- Mewis, I.; Appel, H.M.; Hom, A.; Raina, R.; Schultz, J.C. Major signaling pathways modulate arabidopsis glucosinolate accumulation and response to both phloem-feeding and chewing insects. Plant Physiol. 2005, 138, 1149–1162. [Google Scholar] [CrossRef] [PubMed]
- Kliebenstein, D.; Pedersen, D.; Barker, B.; Mitchell-Olds, T. Comparative analysis of quantitative trait loci controlling glucosinolates, myrosinase and insect resistance in Arabidopsis thaliana. Genetics 2002, 161, 325–332. [Google Scholar] [PubMed]
- Mikkelsen, M.D.; Petersen, B.L.; Glawischnig, E.; Jensen, A.B.; Andreasson, E.; Halkier, B.A. Modulation of CYP79 genes and glucosinolate profiles in arabidopsis by defense signaling pathways. Plant Physiol. 2003, 131, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Mithen, R.F.; Magrath, R. Glucosinolates and resistance to Leptosphaeria maculans in wild and cultivated Brassica species. Plant Breeding 1992, 108, 60–68. [Google Scholar] [CrossRef]
- Sexton, A.C.; Kirkegaard, J.A.; Howlett, B.J. Glucosinolates in Brassica juncea and resistance to australian isolates of Leptosphaeria maculans, the blackleg fungus. Australasian Plant Pathol. 1999, 28, 95–102. [Google Scholar] [CrossRef]
- Li, Y.; Kiddle, G.; Bennett, R.; Wallsgrove, R. Local and systemic changes in glucosinolates in chinese and european cultivars of oilseed rape (Brassica napus L.) after inoculation with Sclerotinia sclerotiorum (stem rot). Ann. Appl. Biol. 1999, 134, 45–58. [Google Scholar] [CrossRef]
- Sotelo, T.; Lema, M.; Soengas, P.; Cartea, M.; Velasco, P. In vitro activity of glucosinolates and their degradation products against Brassica pathogenic bacteria and fungi. Appl. Environ. Microbiol. 2014, AEM, 03142–03144. [Google Scholar]
- Giamoustaris, A.; Mithen, R. Glucosinolates and disease resistance in oilseed rape (Brassica napus ssp. oleifera). Plant Pathol. 1997, 46, 271–275. [Google Scholar] [CrossRef]
- Doughty, K.J.; Porter, A.J.R.; Morton, A.M.; Kiddle, G.; Bock, C.H.; Wallsgrove, R. Variation in the glucosinolate content of oilseed rape (Brassica napus L.) leaves. Ann. Appl. Biol. 1991, 118, 469–477. [Google Scholar] [CrossRef]
- Ludwig-Müller, J.; Bennett, R.; Kiddle, G.; Ihmig, S.; Ruppel, M.; Hilgenberg, W. The host range of Plasmodiophora brassicae and its relationship to endogenous glucosinolate content. New Phytol. 1999, 141, 443–458. [Google Scholar] [CrossRef]
- Brader, G.; Mikkelsen, M.D.; Halkier, B.A.; Tapio Palva, E. Altering glucosinolate profiles modulates disease resistance in plants. Plant J. 2006, 46, 758–767. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez-Vallet, A.; Ramos, B.; Bednarek, P.; López, G.; Piślewska-Bednarek, M.; Schulze-Lefert, P.; Molina, A. Tryptophan-derived secondary metabolites in Arabidopsis thaliana confer non-host resistance to necrotrophic Plectosphaerella cucumerina fungi. Plant J. 2010, 63, 115–127. [Google Scholar] [CrossRef]
- Buxdorf, K.; Yaffe, H.; Barda, O.; Levy, M. The effects of glucosinolates and their breakdown products on necrotrophic fungi. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Bednarek, P.; Piślewska-Bednarek, M.; Svatoš, A.; Schneider, B.; Doubský, J.; Mansurova, M.; Humphry, M.; Consonni, C.; Panstruga, R.; Sanchez-Vallet, A.; et al. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 2009, 323, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Hiruma, K.; Fukunaga, S.; Bednarek, P.; Piślewska-Bednarek, M.; Watanabe, S.; Narusaka, Y.; Shirasu, K.; Takano, Y. Glutathione and tryptophan metabolism are required for Arabidopsis immunity during the hypersensitive response to hemibiotrophs. Proc. Natl. Acad. Sci. USA 2013, 110, 9589–9594. [Google Scholar] [CrossRef] [PubMed]
- Yi, G.-E.; Robin, A.H.K.; Yang, K.; Park, J.-I.; Hwang, B.H.; Nou, I.-S. Exogenous methyl jasmonate and salicylic acid induce subspecies-specific patterns of glucosinolate accumulation and gene expression in Brassica oleracea L. Molecules 2016, 21, 1417. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Hu, Z.; Zhang, Y.; Li, Y.; Zhou, S.; Chen, G. A putative functional myb transcription factor induced by low temperature regulates anthocyanin biosynthesis in purple kale (brassica oleracea var. Acephala f. Tricolor). Plant Cell Rep. 2012, 31, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Frerigmann, H.; Piślewska-Bednarek, M.; Sánchez-Vallet, A.; Molina, A.; Glawischnig, E.; Gigolashvili, T.; Bednarek, P. Regulation of pathogen-triggered tryptophan metabolism in Arabidopsis thaliana by MYB transcription factors and indole glucosinolate conversion products. Mol. Plant 2016, 9, 682–695. [Google Scholar] [CrossRef]
- Wu, J.; Zhao, Q.; Yang, Q.; Liu, H.; Li, Q.; Yi, X.; Cheng, Y.; Guo, L.; Fan, C.; Zhou, Y. Comparative transcriptomic analysis uncovers the complex genetic network for resistance to Sclerotinia sclerotiorum in Brassica napus. Sci. Rep. 2016, 6, 1–15. [Google Scholar] [CrossRef]
- Gigolashvili, T.; Yatusevich, R.; Berger, B.; Müller, C.; Flügge, U.I. The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007, 51, 247–261. [Google Scholar] [CrossRef]
- Frerigmann, H.; Gigolashvili, T. MYB34, MYB51, and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol. Plant 2014, 7, 814–828. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Rahman, M.H.; Liang, Y.; Shah, S.; Kav, N.N. Characterization of defense signaling pathways of Brassica napus and Brassica carinata in response to Sclerotinia sclerotiorum challenge. Plant Mol. Biol. Rep. 2010, 28, 253–263. [Google Scholar] [CrossRef]
- Sticher, L.; Mauch-Mani, B.; Métraux, J.P. Systemic acquired resistance. Ann. Rev. Phytopathol. 1997, 35, 235–270. [Google Scholar] [CrossRef] [PubMed]
- Durrant, W.E.; Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 2004, 42, 185–209. [Google Scholar] [CrossRef] [PubMed]
- Grant, M.; Lamb, C. Systemic immunity. Curr. Opin. Plant Biol. 2006, 9, 414–420. [Google Scholar] [CrossRef] [PubMed]
- Alkooranee, J.T.; Aledan, T.R.; Xiang, J.; Lu, G.; Li, M. Induced systemic resistance in two genotypes of Brassica napus (AACC) and Raphanus oleracea (RRCC) by Trichoderma isolates against Sclerotinia sclerotiorum. Am. J. Plant Sci. 2015, 6, 1662. [Google Scholar] [CrossRef]
- Alkooranee, J.T.; Yin, Y.; Aledan, T.R.; Jiang, Y.; Lu, G.; Wu, J.; Li, M. Systemic resistance to powdery mildew in Brassica napus (aacc) and Raphanus alboglabra (rrcc) by Trichoderma harzianum th12. PLoS ONE 2015, 10, e0142177. [Google Scholar] [CrossRef]
- Sotelo, T.; Velasco, P.; Soengas, P.; Rodríguez, V.M.; Cartea, M.E. Modification of leaf glucosinolate contents in Brassica oleracea by divergent selection and effect on expression of genes controlling glucosinolate pathway. Front. Plant Sci. 2016, 7, 1–12. [Google Scholar] [CrossRef]
- Mithen, R.F.; Lewis, B.G.; Fenwick, G.R. In vitro activity of glucosinolates and their products against Leptosphaeria maculans. Trans. Br. Mycol. Soc. 1986, 87, 433–440. [Google Scholar] [CrossRef]
- Xu, L.; Yang, H.; Ren, L.; Chen, W.; Liu, L.; Liu, F.; Zeng, L.; Yan, R.; Chen, K.; Fang, X. Jasmonic acid-mediated aliphatic glucosinolate metabolism is involved in clubroot disease development in Brassica napus L. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Harikrishnan, R.; Del Río, L. Influence of temperature, relative humidity, ascospore concentration, and length of drying of colonized dry bean flowers on white mold development. Plant Dis. 2006, 90, 946–950. [Google Scholar] [CrossRef]
- Godoy, G.; Steadman, J.; Dickman, M.; Dam, R. Use of mutants to demonstrate the role of oxalic acid in pathogenicity of Sclerotinia sclerotiorum on Phaseolus vulgaris. Physiol. Mol. Plant Pathol. 1990, 37, 179–191. [Google Scholar] [CrossRef]
- Zhao, J.; Meng, J. Genetic analysis of loci associated with partial resistance to Sclerotinia sclerotiorum in rapeseed (Brassica napus L.). Theor. Appl. Genet. 2003, 106, 759–764. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Liu, P.; Hong, D.; He, Q.; Yang, G. Improvement of Sclerotinia resistance of a polima cms restorer line of rapeseed via phenotypic selection, marker-assisted background selection and microspore culture. Plant Breeding 2010, 129, 39–44. [Google Scholar] [CrossRef]
- Yi, G.-E.; Robin, A.H.K.; Yang, K.; Park, J.-I.; Kang, J.-G.; Yang, T.-J.; Nou, I.-S. Identification and expression analysis of glucosinolate biosynthetic genes and estimation of glucosinolate contents in edible organs of Brassica oleracea subspecies. Molecules 2015, 20, 13089–13111. [Google Scholar] [CrossRef] [PubMed]
- Laila, R.; Robin, A.H.K.; Yang, K.; Park, J.-I.; Suh, M.C.; Kim, J.; Nou, I.-S. Developmental and genotypic variation in leaf wax content and composition, and in expression of wax biosynthetic genes in Brassica oleracea var. Capitata. Front. Plant Sci. 2017, 7, 1972. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− δδct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, Y.; Yang, X.; Tong, C.; Edwards, D.; Parkin, I.A.; Zhao, M.; Ma, J.; Yu, J.; Huang, S. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014, 5, 3930. [Google Scholar] [CrossRef] [PubMed]
- Seo, M.-S.; Kim, J.S. Understanding of MYB transcription factors involved in glucosinolate biosynthesis in Brassicaceae. Molecules 2017, 22, 1549. [Google Scholar]







| Function of Gene Product | Genes Upregulated in R Line | Genes Upregulated in S Line | Genes Upregulated in both R and S Lines | |
|---|---|---|---|---|
| Higher in R Line | Higher in S Line | |||
| Transcription Factor | MYB28-Bol017019 (1, 10.2); MYB34-Bol017062 (1, 3.08) | MYB28-Bol036743 (1, 5.8); MYB29-Bol008849 (1, 51.0); MYB28-Bol036286 (1, 21.6); MYB28-Bol007795 (1, 2.9) | MYB122-Bol026204 (3, 189.1) | |
| Aliphatic Biosynthesis | ST5b-Bol026202 (3, 5.6); ST5c-Bol030757 (3, 68.6) | AOP2-Bo3g052110 (1, 8.4); AOP2-Bo9g006240 (1, 11.4); FMOGS-OX2-Bol010993 (1, 5.5); GSL-OH-Bol033373 (1, 65.3) | ||
| Indole Biosynthesis | ST5a-Bol039395 (3, 25.3); ST5a-Bol026200 (3, 82.9) | CYP81F1-Bol017375 (1, 2.0); CYP81F1-Bol017376 (3, 7.96) | CYP81F4-Bol032712 (3, 21.1); CYP81F2-Bol026044 (3, 54.5); CYP81F2-Bol014239 (3, 871); CYP81F2-Bol012237 (3, 48.3); IGMT1-Bol007029 (3, 24.7); IGMT2-Bol007030 (3, 171) | CYP81F1-Bol028914 (3, 19.6); CYP81F4-Bol032714 (3, 12.3); IGMT1-Bol020663 (3, 652) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abuyusuf, M.; Robin, A.H.K.; Lee, J.-H.; Jung, H.-J.; Kim, H.-T.; Park, J.-I.; Nou, I.-S. Glucosinolate Profiling and Expression Analysis of Glucosinolate Biosynthesis Genes Differentiate White Mold Resistant and Susceptible Cabbage Lines. Int. J. Mol. Sci. 2018, 19, 4037. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19124037
Abuyusuf M, Robin AHK, Lee J-H, Jung H-J, Kim H-T, Park J-I, Nou I-S. Glucosinolate Profiling and Expression Analysis of Glucosinolate Biosynthesis Genes Differentiate White Mold Resistant and Susceptible Cabbage Lines. International Journal of Molecular Sciences. 2018; 19(12):4037. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19124037
Chicago/Turabian StyleAbuyusuf, Md., Arif Hasan Khan Robin, Ji-Hee Lee, Hee-Jeong Jung, Hoy-Taek Kim, Jong-In Park, and Ill-Sup Nou. 2018. "Glucosinolate Profiling and Expression Analysis of Glucosinolate Biosynthesis Genes Differentiate White Mold Resistant and Susceptible Cabbage Lines" International Journal of Molecular Sciences 19, no. 12: 4037. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19124037