Liver Regeneration: Different Sub-Populations of Parenchymal Cells at Play Choreographed by an Injury-Specific Microenvironment
Abstract
:1. Introduction
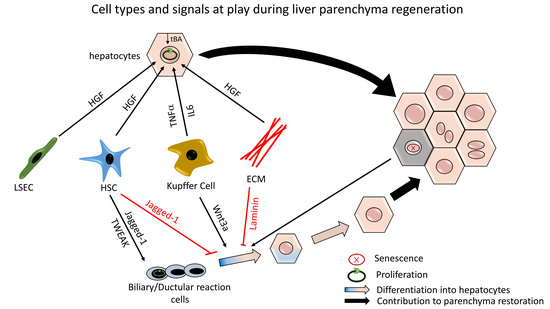
2. Hepatocyte-Mediated Parenchymal Regeneration
2.1. Initiating Molecular Signals
2.2. Terminating Molecular Signals
2.3. Hepatocytes Zonation within the Lobule: Not Only for Metabolism
2.3.1. Pericentral Hepatocytes Expressing Axin2
2.3.2. Periportal Hepatocytes Expressing Sox9 or Mfsd2a
2.3.3. Hepatocytes with High Expression of Telomerase Reverse Transcriptase
2.3.4. Hepatocytes Expressing Lgr4
3. Ductular Reaction-Driven Parenchymal Regeneration in Chronically Injured Liver
3.1. DR Cells Can Differentiate into Hepatocytes
3.2. Molecular Signals Involved in DR Expansion and Differentiation
3.3. Significance of DR-Mediated Parenchymal Regeneration
4. Boosting Hepatocyte or DR-Mediated Parenchymal Regeneration as a Potential Alternative Strategy to Alleviate Liver Insufficiency
4.1. Stimulate in vivo DR-Mediated Parenchymal Regeneration?
4.2. Pitfalls of Liver Regenerating Therapies
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Rappaport, A.M.; Borowy, Z.J.; Lougheed, W.M.; Lotto, N. Subdivisions of Hexagonal Liver Lobules into a Structural and Functional Unit. Anat. Rec. 1954, 119, 11–33. [Google Scholar] [CrossRef]
- Treyer, A.; Mush, A. Hepatocyte Polarity. Compr. Physiol. 2013, 3, 243–287. [Google Scholar]
- Tietz, P.S.; Nicholas, F. Cholangiocyte biology. Curr. Opin. Gastroenterol. 2006, 22, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Theise, N.D.; Saxena, R.; Portmann, B.C.; Thung, S.N.; Yee, H.; Chiriboga, L.; Kumar, A.; Crawford, J.M. The canals of Hering and hepatic stem cells in humans. Hepatology 1999, 30, 1425–1433. [Google Scholar] [CrossRef] [PubMed]
- Saxena, R.; Theise, N. Canals of Hering: Recent insights and current knowledge. Semin. Liver Dis. 2004, 24, 43–48. [Google Scholar]
- Krenkel, O.; Tacke, F. Liver macrophages in tissue homeostasis and disease. Nat. Rev. Immunol. 2017, 17, 306–321. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, T.; Friedman, S.L. Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Michalopoulos, G.K. Hepatostat: Liver regeneration and normal liver tissue maintenance. Hepatology 2017, 65, 1384–1392. [Google Scholar] [CrossRef]
- Malato, Y.; Naqvi, S.; Schürmann, N.; Ng, R.; Wang, B.; Zape, J.; Kay, M.A.; Grimm, D.; Willenbring, H. Fate tracing of mature hepatocytes in mouse liver homeostasis and regeneration. J. Clin. Investig. 2011, 121, 4850–4860. [Google Scholar] [CrossRef] [Green Version]
- Schaub, J.R.; Malato, Y.; Gormond, C.; Willenbring, H. Evidence against a stem cell origin of new hepatocytes in a common mouse model of chronic liver injury. Cell Rep. 2014, 8, 933–939. [Google Scholar] [CrossRef]
- Yanger, K.; Knigin, D.; Zong, Y.; Maggs, L.; Gu, G.; Akiyama, H.; Pikarsky, E.; Stanger, B.Z. Adult hepatocytes are generated by self-duplication rather than stem cell differentiation. Cell Stem Cell 2014, 15, 340–349. [Google Scholar] [CrossRef] [PubMed]
- Michalopoulos, G.K.; DeFrances, M.C.; Ingelmo-Torres, M.; Nixon, S.J.; Ferguson, C.; Kurzchalia, T.; Tebar, F.; Enrich, C.; Parton, R.G.; Pol, A. Liver regeneration. Science 1997, 276, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Fausto, N. Messenger RNA in regenerating liver: Implications for the understanding of regulated growth. Mol. Cell. Biochem. 1984, 59, 131–147. [Google Scholar] [CrossRef] [PubMed]
- Taub, R. Transcriptional control of liver regeneration. FASEB J. 1996, 10, 413–427. [Google Scholar] [CrossRef] [PubMed]
- Michalopoulos, G.K. Liver regeneration. J. Cell. Physiol. 2007, 213, 286–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Wang, X.; Xie, G.; Wang, L.; Hill, C.K.; Deleve, L.D. Liver sinusoidal endothelial cell progenitor cells promote liver regeneration in rats. J. Clin. Investig. 2012, 122, 1567–1573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeishi, T.; Hirano, K.; Kobayashi, T.; Hasegawa, G.; Hatakeyama, K.; Naito, M. The Role of Kupffer Cells in Liver Regeneration. Arch. Histol. Cytol. 1999, 62, 413–422. [Google Scholar] [CrossRef]
- Yamada, Y.; Fausto, N. Deficient liver regeneration after carbon tetrachloride injury in mice lacking type 1 but not type 2 tumor necrosis factor receptor. Am. J. Pathol. 1998, 152, 1577–1589. [Google Scholar]
- Cressman, D.E.; Greenbaum, L.E.; DeAngelis, R.A.; Ciliberto, G.; Furth, E.E.; Poli, V.; Taub, R. Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science 1996, 274, 1379–1383. [Google Scholar] [CrossRef]
- Galun, E.; Zeira, E.; Pappo, O.; Peters, M.; Rose-John, S. Liver regeneration induced by a designer human IL-6/sIL-6R fusion protein reverses severe hepatocellular. FASEB J. 2000, 14, 1979–1987. [Google Scholar] [CrossRef]
- Monga, S.P.S.; Pediaditakis, P.; Mule, K.; Stolz, D.B.; Michalopoulos, G.K. Changes in wnt/β-catenin pathway during regulated growth in rat liver regeneration. Hepatology 2001, 33, 1098–1109. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Behari, J.; Cieply, B.; Michalopoulos, G.K.; Monga, S.P.S. Conditional Deletion of β-Catenin Reveals Its Role in Liver Growth and Regeneration. Gastroenterology 2006, 131, 1561–1572. [Google Scholar] [CrossRef] [PubMed]
- Sekine, S.; Lan, B.Y.A.; Bedolli, M.; Feng, S.; Hebrok, M. Liver-specific loss of beta-catenin blocks glutamine synthesis pathway activity and cytochrome P450 expression in mice. Hepatology 2006, 43, 817–825. [Google Scholar] [CrossRef] [PubMed]
- Croquelois, A.; Blindenbacher, A.; Terracciano, L.; Wang, X.; Langer, I.; Radtke, F.; Heim, M.H. Inducible inactivation of Notch1 causes nodular regenerative hyperplasia in mice. Hepatology 2005, 41, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Keitel, V.; Kubitz, R.; Häussinger, D. Endocrine and paracrine role of bile acids. World J. Gastroenterol. 2008, 14, 5620–5629. [Google Scholar] [CrossRef] [PubMed]
- Borude, P.; Edwards, G.; Walesky, C.; Li, F.; Ma, X.; Kong, B.; Guo, G.L.; Apte, U. Hepatocyte-specific deletion of farnesoid X receptor delays but does not inhibit liver regeneration after partial hepatectomy in mice. Hepatology 2012, 56, 2344–2352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, W.; Ma, K.; Zhang, J.; Qatanani, M.; Cuvillier, J.; Liu, J.; Dong, B.; Huang, X.; Moore, D.D. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science 2006, 312, 233–236. [Google Scholar] [CrossRef] [PubMed]
- Meng, Z.; Wang, Y.; Wang, L.; Jin, W.; Liu, N.; Pan, H.; Liu, L.; Wagman, L.; Forman, B.M.; Huang, W. FXR Regulates Liver Repair after CCl 4 -Induced Toxic Injury. Mol. Endocrinol. 2010, 24, 886–897. [Google Scholar] [CrossRef] [PubMed]
- Jourdainne, V.; Péan, N.; Doignon, I.; Humbert, L.; Rainteau, D.; Tordjmann, T. The bile acid receptor TGR5 and liver regeneration. Dig. Dis. 2015, 33, 319–326. [Google Scholar] [CrossRef]
- Péan, N.; Doignon, I.; Garcin, I.; Besnard, A.; Julien, B.; Liu, B.; Branchereau, S.; Spraul, A.; Guettier, C.; Humbert, L.; et al. The receptor TGR5 protects the liver from bile acid overload during liver regeneration in mice. Hepatology 2013, 58, 1451–1460. [Google Scholar] [CrossRef] [Green Version]
- Frumento, D. Gut Microbiota Role in Liver Regeneration : Evidences and Novel Insights. Innov. Tissue Eng. Regen. Med. 2018, 1, 13–15. [Google Scholar]
- Liu, H.X.; Keane, R.; Sheng, L.; Wan, Y.J.Y. Implications of microbiota and bile acid in liver injury and regeneration. J. Hepatol. 2015, 63, 1502–1510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.X.; Rocha, C.S.; Dandekar, S.; Yvonne Wan, Y.J. Functional analysis of the relationship between intestinal microbiota and the expression of hepatic genes and pathways during the course of liver regeneration. J. Hepatol. 2016, 64, 641–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyaoka, Y.; Ebato, K.; Kato, H.; Arakawa, S.; Shimizu, S.; Miyajima, A. Hypertrophy and Unconventional Cell Division of Hepatocytes Underlie Liver Regeneration. Curr. Biol. 2012, 22, 1166–1175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gentric, G.; Desdouets, C. Polyploidization in liver tissue. Am. J. Pathol. 2014, 184, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Block, G.; Locker, J.; Bowen, W.C.; Petersen, B.E.; Katyal, S.; Strom, S.C.; Howard, A.; Michalopoulos, G.K. Population Expansion, Clonal Growth, and Specific Differentiation Patterns in Primary Cultures of Hepatocytes Induced by HGF/SF, EGF and TGFoL in a Chemically Defined (HGM) Medium. J. Cell Biol. 1996, 132, 1133–1149. [Google Scholar] [CrossRef]
- Rana, B.; Mischoulon, D.; Xie, Y.; Bucher, N.L.; Farmer, S.R. Cell-extracellular matrix interactions can regulate the switch between growth and differentiation in rat hepatocytes: Reciprocal expression of C/EBP alpha and immediate-early growth response transcription factors. Mol. Cell. Biol. 1994, 14, 5858–5869. [Google Scholar] [CrossRef]
- Gkretsi, V.; Bowen, W.C.; Yang, Y.; Wu, C.; Michalopoulos, G.K. Integrin-linked kinase is involved in matrix-induced hepatocyte differentiation. Biochem. Biophys. Res. Commun. 2007, 353, 638–643. [Google Scholar] [CrossRef] [Green Version]
- Cano-Gauci, D.F.; Song, H.H.; Yang, H.; McKerlie, C.; Choo, B.; Shi, W.; Pullano, R.; Piscione, T.D.; Grisaru, S.; Soon, S.; et al. Glypican-3-deficient mice exhibit developmental overgrowth and some of the abnormalities typical of Simpson-Golabi-Behmel syndrome. J. Cell Biol. 1999, 146, 255–264. [Google Scholar] [CrossRef]
- Zhou, D.; Conrad, C.; Xia, F.; Park, J.S.; Payer, B.; Yin, Y.; Lauwers, G.Y.; Thasler, W.; Lee, J.T.; Avruch, J.; et al. Mst1 and Mst2 Maintain Hepatocyte Quiescence and Suppress Hepatocellular Carcinoma Development through Inactivation of the Yap1 Oncogene. Cancer Cell 2009, 16, 425–438. [Google Scholar] [CrossRef]
- Lu, L.; Li, Y.; Kim, S.M.; Bossuyt, W.; Liu, P.; Qiu, Q.; Wang, Y.; Halder, G.; Finegold, M.J.; Lee, J.-S.; et al. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc. Natl. Acad. Sci. USA 2010, 107, 1437–1442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomital, Y. Inhibitory Effect of Transforming Growth Factor-pan DNA Synthesis of Adult Rat Hepatocytes in Primary Culture. Biochem. Biophys. Res. Commun. 1985, 133, 1042–1050. [Google Scholar]
- Bissell, D.M.; Wang, S.S.; Jarnagin, W.R.; Roll, F.J. Cell-specific expression of transforming growth factor-beta in rat liver. Evidence for autocrine regulation of hepatocyte proliferation. J. Clin. Investig. 1995, 96, 447–455. [Google Scholar] [CrossRef]
- Oe, S.; Lemmer, E.R.; Conner, E.A.; Factor, V.M.; Levéen, P.; Larsson, J.; Karlsson, S.; Thorgeirsson, S.S. Intact signaling by transforming growth factor β is not required for termination of liver regeneration in mice. Hepatology 2004, 40, 1098–1105. [Google Scholar] [CrossRef] [Green Version]
- Jungermann, K.; Keitzmann, T. Zonation of Parenchymal and Nonparenchymal Metabolism in Liver. Annu. Rev. Nutr. 1996, 16, 179–203. [Google Scholar] [CrossRef] [PubMed]
- Zajicek, G.; Oren, R.; Weinreb, M. The streaming liver. Liver 1985, 5, 293–300. [Google Scholar] [CrossRef] [PubMed]
- Lorup, C. An autoradiographic study of the 3H-Uridine and 3H-Thymidine incorporation in the regenerating mouse liver. Cell Tissue Kinet. 1977, 10, 477–485. [Google Scholar]
- Rabes, H. Kinetics of hepatocellular proliferation after partial resection of the liver. Prog. Liver Dis. 1976, 5, 83–99. [Google Scholar]
- Bralet, M.P.; Branchereau, S.; Brechot, C.; Ferry, N. Cell lineage study in the liver using retroviral mediated gene transfer. Evidence against the streaming of hepatocytes in normal liver. Am. J. Pathol. 1994, 144, 896–905. [Google Scholar]
- Wang, B.; Zhao, L.; Fish, M.; Logan, C.Y.; Nusse, R. Self-renewing diploid Axin2+ cells fuel homeostatic renewal of the liver. Nature 2015, 524, 180–185. [Google Scholar] [CrossRef] [Green Version]
- Planas-Paz, L.; Orsini, V.; Boulter, L.; Calabrese, D.; Pikiolek, M.; Nigsch, F.; Xie, Y.; Roma, G.; Donovan, A.; Marti, P.; et al. The RSPO–LGR4/5–ZNRF3/RNF43 module controls liver zonation and size. Nat. Cell Biol. 2016, 18, 467–479. [Google Scholar] [CrossRef]
- MacDonald, R.A. “ Lifespan ” of Liver Cells. Arch. Intern. Med. 1961, 107, 335–343. [Google Scholar] [CrossRef]
- Torre, C.; Perret, C.; Colnot, S. Transcription dynamics in a physiological process: β-Catenin signaling directs liver metabolic zonation. Int. J. Biochem. Cell Biol. 2011, 43, 271–278. [Google Scholar] [CrossRef]
- Font-Burgada, J.; Shalapour, S.; Ramaswamy, S.; Hsueh, B.; Rossell, D.; Umemura, A.; Taniguchi, K.; Nakagawa, H.; Valasek, M.A.; Ye, L.; et al. Hybrid Periportal Hepatocytes Regenerate the Injured Liver without Giving Rise to Cancer. Cell 2015, 162, 766–779. [Google Scholar] [CrossRef] [Green Version]
- Pu, W.; Zhang, H.; Huang, X.; Tian, X.; He, L.; Wang, Y.; Zhang, L.; Liu, Q.; Li, Y.; Li, Y.; et al. Mfsd2a+ hepatocytes repopulate the liver during injury and regeneration. Nat. Commun. 2016, 7, 13369. [Google Scholar] [CrossRef]
- Carpentier, R.; Espanol-Suner, R.; Van Hul, N.; Kopp, J.L.; Beaudry, J.; Cordi, S.; Antoniou, A.; Raynaud, P.; Lepreux, S.; Jacquemin, P.; et al. Embryonic ductal plate cells give rise to cholangiocytes, periportal hepatocytes, and adult liver progenitor cells. Gastroenterology 2011, 141, 1432–1438. [Google Scholar] [CrossRef]
- Lin, S.; Nascimento, E.M.; Gajera, C.R.; Chen, L.; Neuhöfer, P.; Garbuzov, A.; Wang, S.; Artandi, S.E. Distributed hepatocytes expressing telomerase repopulate the liver in homeostasis and injury. Nature 2018, 556, 244–248. [Google Scholar] [CrossRef]
- Clerbaux, L.A.; Manco, R.; Leclercq, I. Upstream regulators of hepatic Wnt/β-catenin activity control liver metabolic zonation, development, and regeneration. Hepatology 2016, 64, 1361–1363. [Google Scholar] [CrossRef]
- Lee, V.M.; Cameron, R.G.; Archer, M.C. Zonal location of compensatory hepatocyte proliferation following chemically induced hepatotoxicity in rats and humans. Toxicol. Pathol. 1998, 26, 621–627. [Google Scholar] [CrossRef]
- Drasdo, D.; Hoehme, S.; Hengstler, J.G. How predictive quantitative modelling of tissue organization can inform liver disease pathogenesis. J. Hepatol. 2014, 61, 951–956. [Google Scholar] [CrossRef]
- Halpern, K.B.; Shenhav, R.; Matcovitch-Natan, O.; Tóth, B.; Lemze, D.; Golan, M.; Massasa, E.E.; Baydatch, S.; Landen, S.; Moor, A.E.; et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 2017, 542, 352–357. [Google Scholar] [CrossRef]
- Wiemann, S.U.; Satyanarayana, A.; Tsahuridu, M.; Tillmann, H.L.; Zender, L.; Klempnauer, J.; Flemming, P.; Franco, S.; Blasco, M.A.; Manns, M.P.; et al. Hepatocyte telomere shortening and senescence are general markers of human liver cirrhosis. FASEB J. 2002, 16, 935–942. [Google Scholar] [CrossRef]
- Marshall, A.; Rushbrook, S.; Davies, S.E.; Morris, L.S.; Scott, I.S.; Vowler, S.L.; Coleman, N.; Alexander, G. Relation between hepatocyte G1 arrest, impaired hepatic regeneration, and fibrosis in chronic hepatitis C virus infection. Gastroenterology 2005, 128, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Clerbaux, L.-A.; Van Hul, N.; Gouw, A.S.H.; Manco, R.; Español-Suñer, R.; Leclercq, I.A. Relevance of the CDE and DDC Mouse Models to Study Ductular Reaction in Chronic Human Liver Diseases. In Experimental Animal Models of Human Diseases—An Effective Therapeutic Strategy; In Tech: London, UK, 2017. [Google Scholar] [CrossRef]
- Yasui, O.; Miura, N.; Terada, K.; Kawarada, Y.; Koyama, K.; Sugiyama, T. Isolation of oval cells from Long-Evans Cinnamon rats and their transformation into hepatocytes in vivo in the rat liver. Hepatology 1997, 25, 329–334. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.-Y.; Bird, T.G.; Boulter, L.; Tsuchiya, A.; Cole, A.M.; Hay, T.; Guest, R.V.; Wojtacha, D.; Man, T.Y.; Mackinnon, A.; et al. Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat. Cell Biol. 2015, 17, 971–983. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falkowski, O.; An, H.J.; Ianus, I.A.; Chiriboga, L.; Yee, H.; West, A.B.; Theise, N.D. Regeneration of hepatocyte “buds” in cirrhosis from intrabiliary stem cells. J. Hepatol. 2003, 39, 357–364. [Google Scholar] [CrossRef]
- Roskams, T.A.; Libbrecht, L.; Desmet, V.J. Progenitor cells in diseased human liver. Semin. Liver Dis. 2003, 23, 385–396. [Google Scholar]
- Roskams, T.A.; Theise, N.D.; Balabaud, C.; Bhagat, G.; Bhathal, P.S.; Bioulac-sage, P.; Brunt, E.M.; Crawford, J.M.; Crosby, H.A.; Desmet, V.; et al. Nomenclature of the Finer Branches of the Biliary. Hepatology 2004, 39, 1739–1745. [Google Scholar] [CrossRef]
- Lin, W.R.; Lim, S.N.; McDonald, S.A.C.; Graham, T.; Wright, V.L.; Peplow, C.L.; Humphries, A.; Kocher, H.M.; Wright, N.A.; Dhillon, A.P.; et al. The histogenesis of regenerative nodules in human liver cirrhosis. Hepatology 2010, 51, 1017–1026. [Google Scholar] [CrossRef] [Green Version]
- Huch, M.; Dorrell, C.; Boj, S.F.; Van Es, J.H.; Li, V.S.W.; Van De Wetering, M.; Sato, T.; Hamer, K.; Sasaki, N.; Finegold, M.J.; et al. In vitro expansion of single Lgr5 + liver stem cells induced by Wnt-driven regeneration. Nature 2013, 494, 247–250. [Google Scholar] [CrossRef]
- Kamiya, A.; Kinoshita, T.; Miyajima, A. Oncostatin M and hepatocyte growth factor induce hepatic maturation via distinct signaling pathways. FEBS Lett. 2001, 492, 90–94. [Google Scholar] [CrossRef] [Green Version]
- Lázaro, C.A.; Rhim, J.A.; Yamada, Y.; Fausto, N. Generation of hepatocytes from oval cell precursors in culture. Cancer Res. 1998, 58, 5514–5522. [Google Scholar] [PubMed]
- Michalopoulos, G.K.; Bowen, W.C.; Mule, K.; Luo, J. Gene Expression Patterns During Formation of Tissue in Hepatic Organoid Cultures. Gene Expr. 2018, 11, 55–75. [Google Scholar] [CrossRef]
- Sekhon, S.S.; Tan, X.; Micsenyi, A.; Bowen, W.C.; Monga, S.P.S. Fibroblast growth factor enriches the embryonic liver cultures for hepatic progenitors. Am. J. Pathol. 2004, 164, 2229–2240. [Google Scholar] [CrossRef]
- Rashid, S.T.; Corbineau, S.; Hannan, N.; Marciniak, S.J.; Miranda, E.; Alexander, G.; Huang-Doran, I.; Griffin, J.; Ahrlund-Richter, L.; Skepper, J.; et al. Modeling inherited metabolic disorders of the liver using human induced pluripotent stem cells. J. Clin. Investig. 2010, 120, 3127–3136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, A.L.; Coulter, S.; Liddle, C.; Wong, A.; Eastham-Anderson, J.; French, D.M.; Peterson, A.S.; Sonoda, J. FGF19 regulates cell proliferation, glucose and bile acid metabolism via FGFR4-dependent and independent pathways. PLoS ONE 2011, 6, e17868. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, H.; Yang, C.; LeBleu, V.S.; Soubasakos, M.A.; Giraldo, M.; Zeisberg, M.; Kalluri, R. BMP-7 functions as a novel hormone to facilitate liver regeneration. FASEB J. 2006, 21, 256–264. [Google Scholar] [CrossRef] [PubMed]
- Huch, M.; Gehart, H.; Van Boxtel, R.; Hamer, K.; Blokzijl, F.; Verstegen, M.M.A.; Ellis, E.; Van Wenum, M.; Fuchs, S.A.; De Ligt, J.; et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 2015, 160, 299–312. [Google Scholar] [CrossRef]
- Español-Suñer, R.; Carpentier, R.; Van Hul, N.; Legry, V.; Achouri, Y.; Cordi, S.; Jacquemin, P.; Lemaigre, F.; Leclercq, I.A. Liver progenitor cells yield functional hepatocytes in response to chronic liver injury in mice. Gastroenterology 2012, 143, 1564.e7–1575.e7. [Google Scholar] [CrossRef]
- Raven, A.; Lu, W.; Man, T.Y.; Ferreira-gonzalez, S.; Duibhir, E.O.; Dwyer, B.J.; Meehan, R.R.; Bogorad, R.; Koteliansky, V.; Kotelevtsev, Y.; et al. Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature 2017, 547, 350–354. [Google Scholar] [CrossRef] [Green Version]
- Deng, X.; Zhang, X.; Li, W.; Feng, R.X.; Li, L.; Yi, G.R.; Zhang, X.N.; Yin, C.; Yu, H.Y.; Zhang, J.P.; et al. Chronic Liver Injury Induces Conversion of Biliary Epithelial Cells into Hepatocytes. Cell Stem Cell 2018, 23, 114.e3–122.e3. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo-Torres, D.; Affo’, S.; Coll, M.; Morales-Ibanez, O.; Millàn, C.; Blaya, D.; Alvarez-Guaita, A.; Rentero, C.; Lozano, J.J.; Maestro, A.M.; et al. The Biliary Epithelium Gives Rise to Liver Progenitor Cells. Hepatology 2014, 60, 1367–1377. [Google Scholar] [CrossRef]
- Jörs, S.; Jeliazkova, P.; Ringelhan, M.; Thalhammer, J.; Dürl, S.; Ferrer, J.; Sander, M.; Heikenwalder, M.; Schmid, R.M.; Siveke, J.T.; et al. Lineage fate of ductular reactions in liver injury and carcinogenesis. J. Clin. Investig. 2015, 125, 2445–2457. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.; Upadhyay, N.; Greenbaum, L.E.; Kaestner, K.H. Ablation of Foxl1-Cre-labeled hepatic progenitor cells and their descendants impairs recovery of mice from liver injury. Gastroenterology 2015, 148, 192.e3–202.e3. [Google Scholar] [CrossRef] [PubMed]
- Tarlow, B.D.; Pelz, C.; Naugler, W.E.; Wakefield, L.; Wilson, E.M.; Finegold, M.J.; Grompe, M. Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell 2014, 15, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Yanger, K.; Zong, Y.; Maggs, L.R.; Shapira, S.N.; Maddipati, R.; Aiello, N.M.; Thung, S.N.; Wells, R.G.; Greenbaum, L.E.; Stanger, B.Z. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev. 2013, 27, 719–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanimizu, N.; Nishikawa, Y.; Ichinohe, N.; Akiyama, H.; Mitaka, T. Sry HMG box protein 9-positive (Sox9+) epithelial cell adhesion molecule-negative (EpCAM-) biphenotypic cells derived from hepatocytes are involved in mouse liver regeneration. J. Biol. Chem. 2014, 289, 7589–7598. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, S.; Suzuki, A. Hepatocytes, rather than cholangiocytes, can be the major source of primitive ductules in the chronically injured mouse liver. Am. J. Pathol. 2014, 184, 1468–1478. [Google Scholar] [CrossRef]
- Kamimoto, K.; Kaneko, K.; Kok, C.Y.Y.; Okada, H.; Miyajima, A.; Itoh, T. Heterogeneity and stochastic growth regulation of biliary epithelial cells dictate dynamic epithelial tissue remodeling. eLife 2016, 5, 1–26. [Google Scholar] [CrossRef]
- Schaub, J.R.; Huppert, K.A.; Kurial, S.N.T.; Hsu, B.Y.; Cast, A.E.; Donnelly, B.; Karns, R.A.; Chen, F.; Rezvani, M.; Luu, H.Y.; et al. De novo formation of the biliary system by TGFβ-mediated hepatocyte transdifferentiation Transdifferentiation. Nature 2018, 557, 247–251. [Google Scholar] [CrossRef]
- Duncan, A.; Dorrell, C.; Grompe, M. Stem Cells and Liver Regeneration. Gastroenterology 2009, 137, 466–481. [Google Scholar] [CrossRef] [Green Version]
- Riehle, K.J.; Dan, Y.Y.; Campbell, J.S.; Fausto, N. New concepts in liver regeneration. J. Gastroenterol. Hepatol. 2011, 26, 203–212. [Google Scholar] [CrossRef] [Green Version]
- Knight, B.B.; Yeoh, G.C.T.; Husk, K.L.; Ly, T.; Abraham, L.J.; Yu, C.; Rhim, J.A.; Fausto, N. Impaired Preneoplastic Changes and Liver Tumor Formation in Tumor Necrosis Factor Receptor Type 1 Knockout Mice. J. Exp. Med. 2000, 192, 1809–1818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, Y.; Kirillova, I.; Peschon, J.J.; Fausto, N. Initiation of liver growth by tumor necrosis factor: Deficient liver regeneration in mice lacking type I tumor necrosis factor receptor. Proc. Natl. Acad. Sci. USA 1997, 94, 1441–1446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakubowski, A.; Ambrose, C.; Parr, M.; Lincecum, J.M.; Wang, M.Z.; Zheng, T.S.; Browning, B.; Michaelson, J.S.; Baestcher, M.; Wang, B.; et al. TWEAK induces liver progenitor cell proliferation. J. Clin. Investig. 2005, 115, 2330–2340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Hul, N.; Lanthier, N.; Espanol-Suner, R.; Abarca Quinones, J.; Van Rooijen, N.; Leclercq, I. Kupffer cells influence parenchymal invasion and phenotypic orientation, but not the proliferation, of liver progenitor cells in a murine model of liver injury. Am. J. Pathol. 2011, 179, 1839–1850. [Google Scholar] [CrossRef]
- Ishikawa, T.; Factor, V.M.; Marquardt, J.U.; Raggi, C.; Seo, D.; Kitade, M.; Conner, E.A.; Thorgeirsson, S.S. Hepatocyte growth factor/c-met signaling is required for stem-cell-mediated liver regeneration in mice. Hepatology 2012, 55, 1215–1226. [Google Scholar] [CrossRef] [Green Version]
- Takase, H.M.; Itoh, T.; Ino, S.; Wang, T.; Koji, T.; Akira, S.; Takikawa, Y.; Miyajima, A. FGF7 is a functional niche signal required for stimulation of adult liver progenitor cells that support liver regeneration. Genes Dev. 2013, 27, 169–181. [Google Scholar] [CrossRef] [Green Version]
- Gu, Y.; Wei, W.; Cheng, Y.; Wan, B.; Ding, X.; Wang, H.; Zhang, Y.; Jin, M. A pivotal role of BEX1 in liver progenitor cell expansion in mice. Stem Cell Res. Ther. 2018, 9, 164. [Google Scholar] [CrossRef]
- Boulter, L.; Govaere, O.; Bird, T.; Radulescu, S.; Ramachandran, P.; Pellicoro, A.; Ridgway, R.A.; Seo, S.S.; Spee, B.; Van Rooijen, N.; et al. Macrophage derived Wnt signalling opposes Notch signalling in a Numb mediated manner to specify HPC fate in chronic liver disease in human and mouse. Nat. Med. 2012, 18, 572–579. [Google Scholar] [CrossRef]
- Lu, J.; Zhou, Y.; Hu, T.; Zhang, H.; Shen, M.; Cheng, P.; Dai, W.; Wang, F.; Chen, K.; Zhang, Y.; et al. Notch Signaling Coordinates Progenitor Cell-Mediated Biliary Regeneration Following Partial Hepatectomy. Sci. Rep. 2016, 6, 22754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yimlamai, D.; Christodoulou, C.; Galli, G.G.; Yanger, K.; Pepe-Mooney, B.; Gurung, B.; Shrestha, K.; Cahan, P.; Stanger, B.Z.; Camargo, F.D. Hippo pathway activity influences liver cell fate. Cell 2014, 157, 1324–1338. [Google Scholar] [CrossRef] [PubMed]
- Swiderska-Syn, M.; Xie, G.; Michelotti, G.A.; Jewell, M.L.; Premont, R.T.; Syn, W.K.; Diehl, A.M. Hedgehog regulates yes-associated protein 1 in regenerating mouse liver. Hepatology 2016, 64, 232–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwon, H.; Song, K.; Han, C.; Chen, W.; Wang, Y.; Dash, S.; Lim, K.; Wu, T. Inhibition of hedgehog signaling ameliorates hepatic inflammation in mice with nonalcoholic fatty liver disease. Hepatology 2016, 63, 1155–1169. [Google Scholar] [CrossRef]
- Omenetti, A.; Choi, S.S.; Michelotti, G.A.; Diehl, A.M. Hedgehog signaling in the liver. J. Hepatol. 2011, 54, 366–373. [Google Scholar] [CrossRef] [PubMed]
- Pozniak, K.N.; Pearen, M.A.; Pereira, T.N.; Kramer, C.S.M.; Kalita-De Croft, P.; Nawaratna, S.K.; Fernandez-Rojo, M.A.; Gobert, G.N.; Tirnitz-Parker, J.E.E.; Olynyk, J.K.; et al. Taurocholate Induces Biliary Differentiation of Liver Progenitor Cells Causing Hepatic Stellate Cell Chemotaxis in the Ductular Reaction: Role in Pediatric Cystic Fibrosis Liver Disease. Am. J. Pathol. 2017, 187, 2744–2757. [Google Scholar] [CrossRef] [PubMed]
- Alpini, G.; Ueno, Y.; Glaser, S.S.; Marzioni, M.; Phinizy, J.L.; Francis, H.; LeSage, G. Bile acid feeding increased proliferative activity and apical bile acid transporter expression in both small and large rat cholangiocytes. Hepatology 2001, 34, 868–876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noll, A.T.R.; Cramer, T.; Olde Damink, S.W.M.; Schaap, F.G. Cholangiocarcinoma, gone without the Wnt? World J. Hepatol. 2016, 8, 1093–1096. [Google Scholar] [CrossRef]
- Rao, Y.P.; Studer, E.J.; Stravitz, R.T.; Gupta, S.; Qiao, L.; Dent, P.; Hylemon, P.B. Activation of the Raf-1/MEK/ERK cascade by bile acids occurs via the epidermal growth factor receptor in primary rat hepatocytes. Hepatology 2002, 35, 307–314. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Foster, M.; Al-Dhalimy, M.; Lagasse, E.; Finegold, M.; Grompe, M. The origin and liver repopulating capacity of murine oval cells. Proc. Natl. Acad. Sci. USA 2003, 100 (Suppl. 1), 11881–11888. [Google Scholar] [CrossRef] [Green Version]
- Duffield, J.S.J.; Forbes, S.J.; Constandinou, C.M.; Clay, S.; Partolina, M.; Vuthoori, S.; Wu, S.; Lang, R.; Iredale, J.P. Selective depletion of macrophages reveals distinct, opposing roles during liver injury and repair. J. Clin. 2005, 115, 56–65. [Google Scholar] [CrossRef] [Green Version]
- Melgar-Lesmes, P.; Edelman, E.R. Monocyte-endothelial cell interactions in the regulation of vascular sprouting and liver regeneration in mouse. J. Hepatol. 2015, 63, 917–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangieri, C.W.; McCartt, J.C.; Strode, M.A.; Lowry, J.E.; Balakrishna, P.M. Perioperative hepatocyte growth factor (HGF) infusions improve hepatic regeneration following portal branch ligation (PBL) in rodents. Surg. Endosc. 2017, 31, 2789–2797. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.M.; Fukui, M.; Wang, Z.; Miao, F.; Karriker, M.J.; Seki, E. Interventional potential of recombinant feline hepatocyte growth factor in a mouse model of non-alcoholic steatohepatitis. Front. Endocrinol. 2018, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Preziosi, M.E.; Monga, S.P. Update on the Mechanisms of Liver Regeneration. Semin. Liver Dis. 2017, 37, 141–151. [Google Scholar] [CrossRef] [PubMed]
- Hegade, V.S.; Speight, R.A.; Etherington, R.E.; Jones, D.E.J. Novel bile acid therapeutics for the treatment of chronic liver diseases. Ther. Adv. Gastroenterol. 2016, 9, 376–391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knight, B.; Lim, R.; Yeoh, G.C.; Olynyk, J.K. Interferon-γ exacerbates liver damage, the hepatic progenitor cell response and fibrosis in a mouse model of chronic liver injury. J. Hepatol. 2007, 47, 826–833. [Google Scholar] [CrossRef]
- Knight, B.; Akhurst, B.; Matthews, V.B.; Ruddell, R.G.; Ramm, G.A.; Abraham, L.J.; Olynyk, J.K.; Yeoh, G.C. Attenuated liver progenitor (oval) cell and fibrogenic responses to the choline deficient, ethionine supplemented diet in the BALB/c inbred strain of mice. J. Hepatol. 2007, 46, 134–141. [Google Scholar] [CrossRef]
- Popov, Y.; Schuppan, D. Targeting liver fibrosis: Strategies for development and validation of antifibrotic therapies. Hepatology 2009, 50, 1294–1306. [Google Scholar] [CrossRef]
- Popov, Y.; Patsenker, E.; Stickel, F.; Zaks, J.; Bhaskar, K.R.; Niedobitek, G.; Kolb, A.; Friess, H.; Schuppan, D. Integrin αvβ6 is a marker of the progression of biliary and portal liver fibrosis and a novel target for antifibrotic therapies. J. Hepatol. 2008, 48, 453–464. [Google Scholar] [CrossRef]
- Peng, Z.W.; Ikenaga, N.; Liu, S.B.; Sverdlov, D.Y.; Vaid, K.A.; Dixit, R.; Weinreb, P.H.; Violette, S.; Sheppard, D.; Schuppan, D.; et al. Integrin αvβ6 critically regulates hepatic progenitor cell function and promotes ductular reaction, fibrosis, and tumorigenesis. Hepatology 2016, 63, 217–232. [Google Scholar] [CrossRef] [PubMed]
- Roskams, T. Liver stem cells and their implication in hepatocellular and cholangiocarcinoma. Oncogene 2006, 25, 3818–3822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sigal, S.H.; Brill, S.; Fiorino, A.S.; Reid, L.M. The liver as a stem cell and lineage system. Am. J. Physiol. 1992, 263, G139–G148. [Google Scholar] [PubMed]
- Mu, X.; Español-suñer, R.; Mederacke, I.; Affò, S.; Manco, R.; Sempoux, C.; Lemaigre, F.P.; Adili, A.; Yuan, D.; Weber, A.; et al. Hepatocellular carcinoma originates from hepatocytes and not from the progenitor/biliary compartment. J. Clin. Investig. 2015, 125, 3891–3903. [Google Scholar] [CrossRef] [PubMed]


© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manco, R.; Leclercq, I.A.; Clerbaux, L.-A. Liver Regeneration: Different Sub-Populations of Parenchymal Cells at Play Choreographed by an Injury-Specific Microenvironment. Int. J. Mol. Sci. 2018, 19, 4115. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19124115
Manco R, Leclercq IA, Clerbaux L-A. Liver Regeneration: Different Sub-Populations of Parenchymal Cells at Play Choreographed by an Injury-Specific Microenvironment. International Journal of Molecular Sciences. 2018; 19(12):4115. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19124115
Chicago/Turabian StyleManco, Rita, Isabelle A. Leclercq, and Laure-Alix Clerbaux. 2018. "Liver Regeneration: Different Sub-Populations of Parenchymal Cells at Play Choreographed by an Injury-Specific Microenvironment" International Journal of Molecular Sciences 19, no. 12: 4115. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19124115