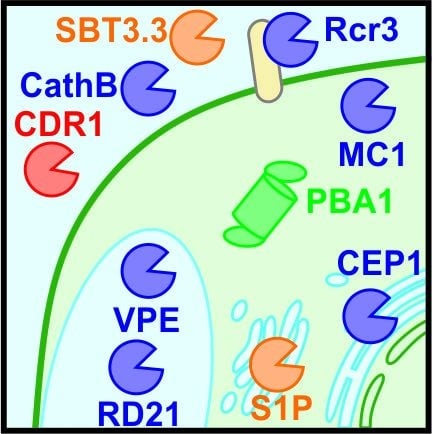
Ten Prominent Host Proteases in Plant-Pathogen Interactions
Abstract
:1. Introduction
2. Apoplastic Proteases
2.1. Subtilase 3.3 (SBT3.3) Regulates the Priming of the Plant Immune Response
2.2. Cathepsin B (CathB) Is a Positive Regulator of Hypersensitive Response (HR)
2.3. Constitutive Disease Resistance-1 (CDR1) Promotes the Release of Systemic Defence Signals
2.4. Cysteine Protease Rcr3 Is a Coreceptor for Perception of Unrelated Pathogens
3. Cytonuclear Proteases
3.1. Arabidopsis thaliana Metacaspase-1 (AtMC1) Is a Positive Regulator of HR
3.2. The Proteasome Is a Positive Regulator of HR
4. Vacuolar Proteases
4.1. Vacuolar Processing Enzymes (VPEs) Regulate Vacuolar Rupture during Virus-Induced HR
4.2. Papain-Like Proteases C14/RD21 Have a Complex Regulation
5. Endomembrane Proteases
5.1. Endoplasmic Reticulum (ER) Resident AtCEP1 Facilitates Fungal Immunity
5.2. Golgi-Localised Site-1-Protease (S1P) Controls Rapid Alkalinisation Factor 23 (RALF23) Peptide Signalling
6. Discussion
Acknowledgments
Conflicts of Interest
References
- Diaz-Mendoza, M.; Velasco-Arroyo, B.; Santamaria, M.E.; González-Melendi, P.; Martinez, M.; Diaz, I. Plant senescence and proteolysis: Two processes with one destiny. Genet. Mol. Biol. 2016, 39, 329–338. [Google Scholar] [CrossRef] [PubMed]
- Ehrmann, M.; Clausen, T. Proteolysis as a regulatory mechanism. Annu. Rev. Genet. 2004, 38, 709–724. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, N.D.; Barrett, A.J.; Finn, R. Twenty years of the MEROPS database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2016, 44. [Google Scholar] [CrossRef] [PubMed]
- Van der Hoorn, R.A.L. Plant proteases: From phenotypes to molecular mechanisms. Annu. Rev. Plant Biol. 2008, 59, 191–223. [Google Scholar] [CrossRef] [PubMed]
- Salvesen, G.S.; Hempel, A.; Coll, N.S. Protease signaling in animal and plant-regulated cell death. FEBS J. 2016, 283, 2577–2598. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, J.; Sousa Silva, M.; Figueiredo, A. Subtilisin-like proteases in plant defence: The past, the present and beyond. Mol. Plant Pathol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Jashni, M.K.; Mehrabi, R.; Collemare, J.; Mesarich, C.H.; de Wit, P.J.G.M. The battle in the apoplast: Further insights into the roles of proteases and their inhibitors in plant-pathogen interactions. Front. Plant Sci. 2015, 6, 584. [Google Scholar] [CrossRef] [PubMed]
- Zipfel, C. Plant pattern-recognition receptors. Trends Immunol. 2014, 35, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Buscaill, P.; Rivas, S. Transcriptional control of plant defence responses. Curr. Opin. Plant Biol. 2014, 20, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Malamy, J.; Carr, J.P.; Klessig, D.F.; Raskin, I. Salicylic acid: A likely endogenous signal in the resistance response of tobacco to viral infection. Science 1990, 250, 1002–1004. [Google Scholar] [CrossRef] [PubMed]
- Vlot, A.C.; Dempsey, D.A.; Klessig, D.F. Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 2009, 47, 177–206. [Google Scholar] [CrossRef] [PubMed]
- Yalpani, N.; Silverman, P.; Wilson, T.M.; Kleier, D.A.; Raskin, I. Salicylic acid is a systemic signal and an inducer of pathogenesis-related proteins in virus-infected tobacco. Plant Cell 1991, 3, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Meng, X.; Shan, L.; He, P. Transcriptional regulation of pattern-triggered immunity in plants. Cell Host Microbe 2016, 19, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Dickman, M.B.; Fluhr, R. Centrality of host cell death in plant-microbe interactions. Annu. Rev. Phytopathol. 2013, 51, 543–570. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, V.; López, A.; Mauch-Mani, B.; Gil, M.J.; Vera, P. An extracellular subtilase switch for immune priming in Arabidopsis. PLoS Pathog. 2013, 9, e1003445. [Google Scholar] [CrossRef] [PubMed]
- McLellan, H.; Gilroy, E.M.; Yun, B.-W.; Birch, P.R.J.; Loake, G.J. Functional redundancy in the Arabidopsis Cathepsin B gene family contributes to basal defence, the hypersensitive response and senescence. New Phytol. 2009, 183, 408–418. [Google Scholar] [CrossRef] [PubMed]
- Gilroy, E.M.; Hein, I.; van der Hoorn, R.; Boevink, P.C.; Venter, E.; McLellan, H.; Kaffarnik, F.; Hrubikova, K.; Shaw, J.; Holeva, M.; et al. Involvement of cathepsin B in the plant disease resistance hypersensitive response. Plant J. 2007, 52, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, Y.; Cai, Y.-M.; Bonneau, L.; Rotari, V.; Danon, A.; McKenzie, E.A.; McLellan, H.; Mach, L.; Gallois, P. Inhibition of cathepsin B by caspase-3 inhibitors blocks programmed cell death in Arabidopsis. Cell Death Differ. 2016, 23, 1493–1501. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.-M.; Yu, J.; Ge, Y.; Mironov, A.; Gallois, P. Two proteases with caspase-3-like activity, cathepsin B and proteasome, antagonistically control ER-stress-induced programmed cell death in Arabidopsis. New Phytol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Suzuki, H.; Borevitz, J.; Blount, J.; Guo, Z.; Patel, K.; Dixon, R.A.; Lamb, C. An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J. 2004, 23, 980–988. [Google Scholar] [CrossRef] [PubMed]
- Prasad, B.D.; Creissen, G.; Lamb, C.; Chattoo, B.B. Overexpression of rice (Oryza sativa L.) OsCDR1 leads to constitutive activation of defense responses in rice and Arabidopsis. Mol. Plant-Microbe Interact. 2009, 22, 1635–1644. [Google Scholar] [CrossRef] [PubMed]
- Lozano-Torres, J.L.; Wilbers, R.H.P.; Gawronski, P.; Boshoven, J.C.; Finkers-Tomczak, A.; Cordewener, J.H.G.; America, A.H.P.; Overmars, H.A.; van ’t Klooster, J.W.; Baranowski, L.; et al. Dual disease resistance mediated by the immune receptor Cf-2 in tomato requires a common virulence target of a fungus and a nematode. Proc. Natl. Acad. Sci. USA 2012, 109, 10119–10124. [Google Scholar] [CrossRef] [PubMed]
- Rooney, H.C.E.; van’t Klooster, J.W.; van der Hoorn, R.A.L.; Joosten, M.H.A.J.; Jones, J.D.G.; de Wit, P.J.G.M. Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 2005, 308, 1783–1786. [Google Scholar] [CrossRef] [PubMed]
- Shabab, M.; Shindo, T.; Gu, C.; Kaschani, F.; Pansuriya, T.; Chintha, R.; Harzen, A.; Colby, T.; Kamoun, S.; van der Hoorn, R.A.L. Fungal effector protein AVR2 targets diversifying defense-related Cys proteases of tomato. Plant Cell 2008, 20, 1169–1183. [Google Scholar] [CrossRef] [PubMed]
- Ilyas, M.; Hörger, A.C.; Bozkurt, T.O.; van den Burg, H.A.; Kaschani, F.; Kaiser, M.; Belhaj, K.; Smoker, M.; Joosten, M.H.A.J.; Kamoun, S.; et al. Functional divergence of two secreted immune proteases of tomato. Curr. Biol. 2015, 25, 2300–2306. [Google Scholar] [CrossRef] [PubMed]
- Coll, N.S.; Vercammen, D.; Smidler, A.; Clover, C.; van Breusegem, F.; Dangl, J.L.; Epple, P. Arabidopsis type I metacaspases control cell death. Science 2010, 330, 1393–1397. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.; Tang, S.; Stallmann, A.; Dangl, J.L.; Bonardi, V. Genetic requirements for signaling from an autoactive plant NB-LRR intracellular innate immune receptor. PLoS Genet. 2013, 9, e1003465. [Google Scholar] [CrossRef] [PubMed]
- Lema Asqui, S.; Vercammen, D.; Serrano, I.; Valls, M.; Rivas, S.; van Breusegem, F.; Conlon, F.L.; Dangl, J.L.; Coll, N.S. AtSERPIN1 is an inhibitor of the metacaspase AtMC1-mediated cell death and autocatalytic processing in planta. New Phytol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Dietrich, R.A.; Delaney, T.P.; Uknes, S.J.; Ward, E.R.; Ryals, J.A.; Dangl, J.L. Arabidopsis mutants simulating disease resistance response. Cell 1994, 77, 565–577. [Google Scholar] [CrossRef]
- Üstün, S.; Sheikh, A.; Gimenez-Ibanez, S.; Jones, A.; Ntoukakis, V.; Börnke, F. The proteasome acts as a hub for plant immunity and is targeted by Pseudomonas type III effectors. Plant Physiol. 2016, 172, 1941–1958. [Google Scholar] [CrossRef] [PubMed]
- Spoel, S.H.; Mou, Z.; Tada, Y.; Spivey, N.W.; Genschik, P.; Dong, X. Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 2009, 137, 860–872. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.Q.; Yan, S.; Saleh, A.; Wang, W.; Ruble, J.; Oka, N.; Mohan, R.; Spoel, S.H.; Tada, Y.; Zheng, N.; et al. NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 2012, 486, 228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Löwe, J.; Stock, D.; Jap, B.; Zwickl, P.; Baumeister, W.; Huber, R. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science 1995, 268, 533–539. [Google Scholar] [CrossRef] [PubMed]
- Kurepa, J.; Smalle, J.A. Structure, function and regulation of plant proteasomes. Biochimie 2008, 90, 324–335. [Google Scholar] [CrossRef] [PubMed]
- Murata, S.; Yashiroda, H.; Tanaka, K. Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 2009, 10, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Hatsugai, N.; Iwasaki, S.; Tamura, K.; Kondo, M.; Fuji, K.; Ogasawara, K.; Nishimura, M.; Hara-Nishimura, I. A novel membrane fusion-mediated plant immunity against bacterial pathogens. Genes Dev. 2009, 23, 2496–2506. [Google Scholar] [CrossRef] [PubMed]
- Del Pozo, O.; Lam, E. Caspases and programmed cell death in the hypersensitive response of plants to pathogens. Curr. Biol. 1998, 8, 1129–1132. [Google Scholar] [CrossRef]
- Bonneau, L.; Ge, Y.; Drury, G.E.; Gallois, P. What happened to plant caspases? J. Exp. Bot. 2008, 59, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Dahan, J.; Etienne, P.; Petitot, A.-S.; Houot, V.; P Blein, J.; Suty, L. Cryptogein affects expression of α3, α6 and β1 20S proteasome subunits encoding genes in tobacco. J. Exp. Bot. 2001, 52, 1947–1948. [Google Scholar] [CrossRef] [PubMed]
- Suty, L.; Lequeu, J.; Lançon, A.; Etienne, P.; Petitot, A.-S.; Blein, J.-P. Preferential induction of 20S proteasome subunits during elicitation of plant defense reactions: Towards the characterization of plant defense proteasomes. Int. J. Biochem. Cell Biol. 2003, 35, 637–650. [Google Scholar] [CrossRef]
- Rojo, E.; Zouhar, J.; Carter, C.; Kovaleva, V.; Raikhel, N. V A unique mechanism for protein processing and degradation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2003, 100, 7389–7394. [Google Scholar] [CrossRef] [PubMed]
- Ito, J.; Fukuda, H. ZEN1 is a key enzyme in the degradation of nuclear DNA during programmed cell death of tracheary elements. Plant Cell 2002, 14, 3201–3211. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.M. Programmed cell death in development and defense. Plant Physiol. 2001, 125, 94–97. [Google Scholar] [CrossRef] [PubMed]
- Hatsugai, N.; Kuroyanagi, M.; Yamada, K.; Meshi, T.; Tsuda, S.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 2004, 305, 855–858. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Chandrasekar, B.; Oeljeklaus, J.; Misas-Villamil, J.C.; Wang, Z.; Shindo, T.; Bogyo, M.; Kaiser, M.; van der Hoorn, R.A.L. Subfamily-specific fluorescent probes for cysteine proteases display dynamic protease activities during seed germination. Plant Physiol. 2015, 168, 1462–1475. [Google Scholar] [CrossRef] [PubMed]
- Hatsugai, N.; Yamada, K.; Goto-Yamada, S.; Hara-Nishimura, I. Vacuolar processing enzyme in plant programmed cell death. Front. Plant Sci. 2015, 6, 234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kabbage, M.; Kessens, R.; Bartholomay, L.C.; Williams, B. The Life and Death of a Plant Cell. Annu. Rev. Plant Biol. 2017, 68, 375–404. [Google Scholar] [CrossRef] [PubMed]
- Kuroyanagi, M.; Yamada, K.; Hatsugai, N.; Kondo, M.; Nishimura, M.; Hara-Nishimura, I. Vacuolar processing enzyme is essential for mycotoxin-induced cell death in Arabidopsis thaliana. J. Biol. Chem. 2005, 280, 32914–32920. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Dong, S.; Wang, M.; Wang, W.; Song, W.; Dou, X.; Zheng, X.; Zhang, Z. The role of vacuolar processing enzyme (VPE) from Nicotiana benthamiana in the elicitor-triggered hypersensitive response and stomatal closure. J. Exp. Bot. 2010, 61, 3799–3812. [Google Scholar] [CrossRef] [PubMed]
- Urquhart, W.; Gunawardena, A.H.L.A.N.; Moeder, W.; Ali, R.; Berkowitz, G.A.; Yoshioka, K. The chimeric cyclic nucleotide-gated ion channel ATCNGC11/12 constitutively induces programmed cell death in a Ca2+ dependent manner. Plant Mol. Biol. 2007, 65, 747–761. [Google Scholar] [CrossRef] [PubMed]
- Pajerowska-Mukhtar, K.; Dong, X. A kiss of death—proteasome-mediated membrane fusion and programmed cell death in plant defense against bacterial infection. Genes Dev. 2009, 23, 2449–2454. [Google Scholar] [CrossRef] [PubMed]
- Shindo, T.; Misas-Villamil, J.C.; Hörger, A.C.; Song, J.; van der Hoorn, R.A.L. A Role in immunity for Arabidopsis cysteine protease RD21, the ortholog of the tomato immune protease C14. PLoS ONE 2012, 7, e29317. [Google Scholar] [CrossRef] [PubMed]
- Yamada, K.; Matsushima, R.; Nishimura, M.; Hara-Nishimura, I. A slow maturation of a cysteine protease with a granulin domain in the vacuoles of senescing Arabidopsis leaves. Plant Physiol. 2001, 127, 1626–1634. [Google Scholar] [CrossRef] [PubMed]
- Bozkurt, T.O.; Schornack, S.; Win, J.; Shindo, T.; Ilyas, M.; Oliva, R.; Cano, L.M.; Jones, A.M.E.; Huitema, E.; van der Hoorn, R.A.L.; et al. Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc. Natl. Acad. Sci. USA 2011, 108, 20832–20837. [Google Scholar] [CrossRef] [PubMed]
- Kaschani, F.; Shabab, M.; Bozkurt, T.; Shindo, T.; Schornack, S.; Gu, C.; Ilyas, M.; Win, J.; Kamoun, S.; van der Hoorn, R.A.L. An effector-targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol. 2010, 154, 1794–1804. [Google Scholar] [CrossRef] [PubMed]
- Shindo, T.; Kaschani, F.; Yang, F.; Kovács, J.; Tian, F.; Kourelis, J.; Hong, T.N.; Colby, T.; Shabab, M.; Chawla, R.; et al. Screen of non-annotated small secreted proteins of Pseudomonas syringae reveals a virulence factor that inhibits tomato immune proteases. PLoS Pathog. 2016, 12, e1005874. [Google Scholar] [CrossRef] [PubMed]
- Lampl, N.; Alkan, N.; Davydov, O.; Fluhr, R. Set-point control of RD21 protease activity by AtSerpin1 controls cell death in Arabidopsis. Plant J. 2013, 74, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Rustgi, S.; Boex-Fontvieille, E.; Reinbothe, C.; von Wettstein, D.; Reinbothe, S. Serpin1 and WSCP differentially regulate the activity of the cysteine protease RD21 during plant development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, 2212–2217. [Google Scholar] [CrossRef] [PubMed]
- Ondzighi, C.A.; Christopher, D.A.; Cho, E.J.; Chang, S.-C.; Staehelin, L.A. Arabidopsis protein disulfide isomerase-5 inhibits cysteine proteases during trafficking to vacuoles before programmed cell death of the endothelium in developing seeds. Plant Cell 2008, 20, 2205–2220. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Shabab, M.; Strasser, R.; Wolters, P.J.; Shindo, T.; Niemer, M.; Kaschani, F.; Mach, L.; van der Hoorn, R.A.L. Post-translational regulation and trafficking of the granulin-containing protease RD21 of Arabidopsis thaliana. PLoS ONE 2012, 7, e32422. [Google Scholar] [CrossRef] [PubMed]
- Kørner, C.J.; Du, X.; Vollmer, M.E.; Pajerowska-Mukhtar, K.M. Endoplasmic reticulum stress signaling in plant immunity—At the crossroad of life and death. Int. J. Mol. Sci. 2015, 16, 26582–26598. [Google Scholar] [CrossRef] [PubMed]
- Eichmann, R.; Schäfer, P. The endoplasmic reticulum in plant immunity and cell death. Front. Plant Sci. 2012, 3, 200. [Google Scholar] [CrossRef] [PubMed]
- Höwing, T.; Huesmann, C.; Hoefle, C.; Nagel, M.-K.; Isono, E.; Hückelhoven, R.; Gietl, C. Endoplasmic reticulum KDEL-tailed cysteine endopeptidase 1 of Arabidopsis (AtCEP1) is involved in pathogen defense. Front. Plant Sci. 2014, 5, 58. [Google Scholar] [CrossRef] [PubMed]
- Höwing, T.; Dann, M.; Hoefle, C.; Hückelhoven, R.; Gietl, C. Involvement of Arabidopsis thaliana endoplasmic reticulum KDEL-tailed cysteine endopeptidase 1 (AtCEP1) in powdery mildew-induced and AtCPR5-controlled cell death. PLoS ONE 2017, 12, e0183870. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Liu, D.; Lv, X.; Wang, Y.; Xun, Z.; Liu, Z.; Li, F.; Lu, H. The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. Plant Cell 2014, 26, 2939–2961. [Google Scholar] [CrossRef] [PubMed]
- Bowling, S.A.; Clarke, J.D.; Liu, Y.; Klessig, D.F.; Dong, X. The cpr5 mutant of Arabidopsis expresses both NPR1-dependent and NPR1-independent resistance. Plant Cell 1997, 9, 1573–1584. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.; Liu, J.-X.; Guo, H.; Yin, Y.; Howell, S.H. Regulation and processing of a plant peptide hormone, AtRALF23, in Arabidopsis. Plant J. 2009, 59, 930–939. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.-X.; Srivastava, R.; Che, P.; Howell, S.H. Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 2007, 51, 897–909. [Google Scholar] [CrossRef] [PubMed]
- Stegmann, M.; Monaghan, J.; Smakowska-Luzan, E.; Rovenich, H.; Lehner, A.; Holton, N.; Belkhadir, Y.; Zipfel, C. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 2017, 355, 287–289. [Google Scholar] [CrossRef] [PubMed]
- Masachis, S.; Segorbe, D.; Turrà, D.; Leon-Ruiz, M.; Fürst, U.; El Ghalid, M.; Leonard, G.; López-Berges, M.S.; Richards, T.A.; Felix, G.; et al. A fungal pathogen secretes plant alkalinizing peptides to increase infection. Nat. Microbiol. 2016, 1, 16043. [Google Scholar] [CrossRef] [PubMed]
- Shalini, S.; Dorstyn, L.; Dawar, S.; Kumar, S. Old, new and emerging functions of caspases. Cell Death Differ. 2015, 22, 526–539. [Google Scholar] [CrossRef] [PubMed]
- McIlwain, D.R.; Berger, T.; Mak, T.W. Caspase functions in cell death and disease. Cold Spring Harb. Perspect. Biol. 2013, 5, a008656. [Google Scholar] [CrossRef] [PubMed]
- Schilling, O.; Huesgen, P.F.; Barré, O.; Auf dem Keller, U.; Overall, C.M. Characterization of the prime and non-prime active site specificities of proteases by proteome-derived peptide libraries and tandem mass spectrometry. Nat. Protoc. 2011, 6, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Coll, N.S.; Smidler, A.; Puigvert, M.; Popa, C.; Valls, M.; Dangl, J.L. The plant metacaspase AtMC1 in pathogen-triggered programmed cell death and aging: Functional linkage with autophagy. Cell Death Differ. 2014, 21, 1399–1408. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Win, J.; Tian, M.; Schornack, S.; Kaschani, F.; Ilyas, M.; van der Hoorn, R.A.L.; Kamoun, S. Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc. Natl. Acad. Sci. USA 2009, 106, 1654–1659. [Google Scholar] [CrossRef] [PubMed]
- Dixon, M.S.; Golstein, C.; Thomas, C.M.; van Der Biezen, E.A.; Jones, J.D. Genetic complexity of pathogen perception by plants: The example of Rcr3, a tomato gene required specifically by Cf-2. Proc. Natl. Acad. Sci. USA 2000, 97, 8807–8814. [Google Scholar] [CrossRef] [PubMed]
- Fortelny, N.; Cox, J.H.; Kappelhoff, R.; Starr, A.E.; Lange, P.F.; Pavlidis, P.; Overall, C.M. Network analyses reveal pervasive functional regulation between proteases in the human protease web. PLoS Biol. 2014, 12, e1001869. [Google Scholar] [CrossRef] [PubMed]
- Grosse-Holz, F.M.; Van der Hoorn, R.A.L. Juggling jobs: Roles and mechanisms of multifunctional protease inhibitors in plants. New Phytol. 2016, 210, 794–807. [Google Scholar] [CrossRef] [PubMed]
- Ghorbani, S.; Hoogewijs, K.; Pečenková, T.; Fernandez, A.; Inzé, A.; Eeckhout, D.; Kawa, D.; De Jaeger, G.; Beeckman, T.; Madder, A.; Van Breusegem, F.; Hilson, P. The SBT6.1 subtilase processes the GOLVEN1 peptide controlling cell elongation. J. Exp. Bot. 2016, 67, 4877–4887. [Google Scholar] [CrossRef] [PubMed]
- Carter, W.C.; Pan, S.; Zouhar, J.; Avila, E.L.; Girke, T.; Raikhel, N.V.; Carter, C.; Pan, S.; Zouhar, J.; Avila, E.L.; et al. The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 2004, 16, 3285–3303. [Google Scholar] [CrossRef] [PubMed]
- Vercammen, D.; Belenghi, B.; van de Cotte, B.; Beunens, T.; Gavigan, J.-A.; De Rycke, R.; Brackenier, A.; Inzé, D.; Harris, J.L.; Van Breusegem, F. Serpin1 of Arabidopsis thaliana is a suicide inhibitor for metacaspase 9. J. Mol. Biol. 2006, 364, 625–636. [Google Scholar] [CrossRef] [PubMed]
- Demir, F.; Niedermaier, S.; Villamor, J.G.; Huesgen, P.F. Quantitative proteomics in plant protease substrate identification. New Phytol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Vizovišek, M.; Vidmar, R.; Fonović, M.; Turk, B. Current trends and challenges in proteomic identification of protease substrates. Biochimie 2016, 122, 77–87. [Google Scholar] [CrossRef] [PubMed]

| Function in Defence | Subcellular Localisation | MEROPS Family | Organism | Known Substrate in Defence? | |
|---|---|---|---|---|---|
| 1. SBT3.3 | Priming | Apoplast | S08, subtilisin-like | A. thaliana | No |
| 2. CathB | Hypersensitive response (HR) | Apoplast (+Vacuole) | C01, papain-like | N. benthamiana, A. thaliana | No |
| 3. CDR1 | Signalling | Apoplast | A01, pepsin-like | A. thaliana | No |
| 4. Rcr3 | Recognition | Apoplast | C01, papain-like | Tomato | No |
| 5. AtMC1 | HR | Cytoplasm (+Nucleus) | C14, metacaspase | A. thaliana | No |
| 6. PBA1 | HR, membrane fusion | Cytoplasm | T01, proteasome | A. thaliana | No |
| 7. VPE | HR, membrane fusion | Vacuole | C13, legumain-like | N. benthamiana, A. thaliana | No |
| 8. C14/RD21 | HR, resistance | Vacuole | C01, papain-like | Tomato, A. thaliana | No |
| 9. CEP1 | Basal resistance | Endoplasmic reticulum (ER) derived compartments | C01, papain-like | A. thaliana | No |
| 10. S1P | Signalling, hormone release | Golgi | S08, subtilisin-like | A. thaliana | RALF23 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thomas, E.L.; Van der Hoorn, R.A.L. Ten Prominent Host Proteases in Plant-Pathogen Interactions. Int. J. Mol. Sci. 2018, 19, 639. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19020639
Thomas EL, Van der Hoorn RAL. Ten Prominent Host Proteases in Plant-Pathogen Interactions. International Journal of Molecular Sciences. 2018; 19(2):639. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19020639
Chicago/Turabian StyleThomas, Emma L., and Renier A. L. Van der Hoorn. 2018. "Ten Prominent Host Proteases in Plant-Pathogen Interactions" International Journal of Molecular Sciences 19, no. 2: 639. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19020639