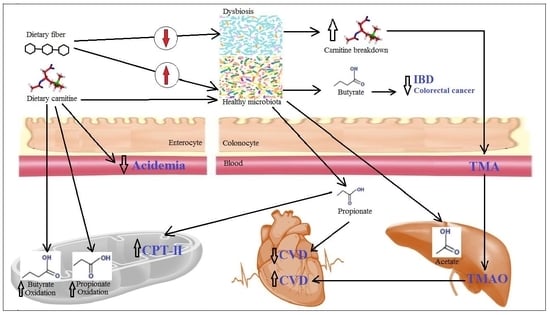
The Impact of Carnitine on Dietary Fiber and Gut Bacteria Metabolism and Their Mutual Interaction in Monogastrics
Abstract
:1. Introduction
2. Carnitine and Bacterial Cell Functions
3. Carnitine and Bacterial Protection
4. Microbial Carnitine Production
5. The Interaction between Carnitine and the Gut Microbial Community
6. Carnitine and Fatty Acid Regulation
7. Fiber–Carnitine Interaction
8. Conclusions
Acknowledgments
Conflicts of Interest
Abbreviations
| ABC | ATP-binding cassette |
| ATP | Adenosine triphosphate |
| BBD | γ-Butyrobetaine dioxygenase |
| BCCT | Betainecholinecarnitine transporter |
| CDH | Carnitine dehydrogenase |
| CPT-II | Carnitine palmitoyltransferase II |
| CSF | Sporulation-stimulating factor |
| CVD | Cardiovascular disease |
| HocS | Hydrolase of O-acylcarnitine, short-chains |
| Hsp27 | Heat-shock inducible protein27 |
| HTML | 3-Hydroxy trimethyl lysine |
| IBD | Inflammatory bowel disease |
| L-CDH | l-Carnitine dehydrogenase |
| LCFAs | Long chain fatty acids |
| LSP | Lipopolysaccharide |
| MCT1 | Monocarboxylate transporter1 |
| NSP | Non-starch polysaccharides |
| Octn2 | Organic cation transporter novel 2 |
| PCC | Propionyl-CoA carboxylase |
| PCCA | Propionyl-CoA carboxylase A |
| PCCB | Propionyl-CoA carboxylase B |
| SCFAs | Short chain fatty acids |
| SMCT1 | Sodium coupled monocarboxylate transporter1 |
| TLR4 | Toll-like receptor 4 |
| TMA | Trimethylamine |
| TML | Trimethyl lysine |
| TMABA | 4-Trimethylaminobutyraldehyde |
| TMAO | Trimethylamine-N-oxide |
| TMLD | Trimethyllysine dioxygenase |
References
- Bernal, V.; Sevilla, Á.; Cánovas, M.; Iborra, J.L. Production of L-carnitine by secondary metabolism of bacteria. Microb. Cell Fact. 2007, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, M.A.; Atherton, H.J.; Dodd, M.S.; Lee, P.; Cochlin, L.E.; Radda, G.K.; Clarke, K.; Tyler, D.J. The Cycling of Acetyl-CoA through acetylcarnitine buffers cardiac substrate supply: A hyperpolarised (13)c magnetic resonance study. Circ. Cardiovasc. Imaging 2012, 5, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Flanagan, J.L.; Simmons, P.A.; Vehige, J.; Willcox, M.D.; Garrett, Q. Role of carnitine in disease. Nutr. Metab. 2010, 7, 30. [Google Scholar] [CrossRef] [PubMed]
- Meadows, J.A.; Wargo, M.J. Carnitine in bacterial physiology and metabolism. Microbiology 2015, 161, 1161–1174. [Google Scholar] [CrossRef] [PubMed]
- Vaz, F.M.; Wanders, R.J. Carnitine biosynthesis in mammals. Biochem. J. 2002, 361 Pt 3, 417–429. [Google Scholar] [CrossRef]
- Bremer, J. Carnitine—Metabolism and functions. Physiol. Rev. 1983, 63, 1420–1480. [Google Scholar] [CrossRef] [PubMed]
- Brass, E.P. Pharmacokinetic considerations for the therapeutic use of carnitine in hemodialysis patients. Clin. Ther. 1995, 17, 176–185. [Google Scholar] [CrossRef]
- Walter, J.; Ley, R. The human gut microbiome: Ecology and recent evolutionary changes. Annu. Rev. Microbiol. 2011, 65, 411–429. [Google Scholar] [CrossRef] [PubMed]
- Holscher, H.D. Dietary fiber and prebiotics and the gastrointestinal microbiota. Gut Microbes 2017, 8, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Che, D.; Qin, G.; Kong, X.; Farouk, M.H. Effects of dietary non-fiber carbohydrates on composition and function of gut microbiome in monogastrics: A Review. Protein Pept. Lett. 2017, 24, 432–441. [Google Scholar] [CrossRef] [PubMed]
- Duncan, S.H.; Belenguer, A.; Holtrop, G.; Johnstone, A.M.; Flint, H.J.; Lobley, G.E. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl. Environ. Microbiol. 2007, 73, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Cummings, J.H.; Pomare, E.W.; Branch, W.J.; Naylor, C.P.; Macfarlane, G.T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 1987, 28, 1221–1227. [Google Scholar] [CrossRef] [PubMed]
- Delzenne, N.M.; Neyrinck, A.M.; Bäckhed, F.; Cani, P.D. Targeting gut microbiota in obesity: Effects of prebiotics and probiotics. Nat. Rev. Endocrinol. 2011, 7, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Csordas, A. Butyrate, aspirin and colorectal cancer. Eur. J. Cancer Prev. 1996, 5, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Zampa, A.; Silvi, S.; Fabiani, R.; Morozzi, G.; Orpianesi, C.; Cresci, A. Effects of different digestible carbohydrates on bile acid metabolism and SCFA production by human gut micro-flora grown in an in vitro semi-continuous culture. Anaerobe 2004, 10, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Charach, G.; Grosskopf, I.; Rabinovich, A.; Shochat, M.; Weintraub, M.; Rabinovich, P. The association of bile acid excretion and atherosclerotic coronary artery disease. Therap. Adv. Gastroenterol. 2011, 4, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Wolever, T.M.; Spadafora, P.J.; Cunnane, S.C.; Pencharz, P.B. Propionate inhibits incorporation of colonic [1,2-13C]acetate into plasma lipids in humans. Am. J. Clin. Nutr. 1995, 61, 1241–1247. [Google Scholar] [CrossRef] [PubMed]
- Scott, K.P.; Duncan, S.H.; Flint, H.J. Dietary fibre and the gut microbiota. Nutr. Bull. 2008, 33, 201–211. [Google Scholar] [CrossRef]
- Bloemen, J.G.; Venema, K.; van de Poll, M.C.; Olde Damink, S.W.; Buurman, W.A.; Dejong, C.H. Short chain fatty acids exchange across the gut and liver in humans measured at surgery. Clin. Nutr. 2009, 28, 657–661. [Google Scholar] [CrossRef] [PubMed]
- Den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.-J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef] [PubMed]
- MacFabe, D.F. Enteric short-chain fatty acids: Microbial messengers of metabolism, mitochondria, and mind: Implications in autism spectrum disorders. Microb. Ecol. Health Dis. 2015, 26, 28177. [Google Scholar] [CrossRef] [PubMed]
- Marciani, P.; Lindi, C.; Marzo, A.; Arrigoni Martelli, E.; Cardace, G.; Esposito, G. l-carnitine and carnitine ester transport in the rat small intestine. Pharmacol. Res. 1991, 23, 157–162. [Google Scholar] [CrossRef]
- Rebouche, C.J. Kinetics, pharmacokinetics, and regulation of l-carnitine and acetyl-l-carnitine metabolism. Ann. N. Y. Acad. Sci. 2004, 1033, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Rebouche, C.J.; Chenard, C.A. Metabolic fate of dietary carnitine in human adults: Identification and quantification of urinary and fecal metabolites. J. Nutr. 1991, 121, 539–546. [Google Scholar] [CrossRef] [PubMed]
- Ussher, J.R.; Lopaschuk, G.D.; Arduini, A. Gut microbiota metabolism of l-carnitine and cardiovascular risk. Atherosclerosis 2013, 231, 456–461. [Google Scholar] [CrossRef] [PubMed]
- Miura-Fraboni, J.; Kleber, H.-P.; Englard, S. Assimilation of γ-butyrobetaine, and d- and l-carnitine by resting cell suspensions of Acinetobacter calcoaceticus and Pseudomonas putida. Arch. Microbiol. 1982, 133, 217–221. [Google Scholar] [CrossRef]
- Unemoto, T.; Hayashi, M.; Miyaki, K.; Hayashi, M. Formation of trimethylamine from dl-carnitine by Serratia marcescens. Biochim. Biophys. Acta 1966, 121, 220–222. [Google Scholar] [CrossRef]
- Zhu, Y.; Jameson, E.; Crosatti, M.; Schafer, H.; Rajakumar, K.; Bugg, T.D.; Chen, Y. Carnitine metabolism to trimethylamine by an unusual Rieske-type oxygenase from human microbiota. Proc. Natl. Acad. Sci. USA 2014, 111, 4268–4273. [Google Scholar] [CrossRef] [PubMed]
- Lindstedt, G.; Lindstedt, S.; Midtvedt, T.; Tofft, M. The formation and degradation of carnitine in Pseudomonas. Biochemistry 1967, 6, 1262–1270. [Google Scholar] [CrossRef]
- Jebbar, M.; Champion, C.; Blanco, C.; Bonnassie, S. Carnitine acts as a compatible solute in Brevibacterium linens. Res. Microbiol. 1998, 149, 211–219. [Google Scholar] [CrossRef]
- Chen, C.; Malek, A.A.; Wargo, M.J.; Hogan, D.A.; Beattie, G.A. The ATP-binding cassette transporter Cbc (choline/betaine/carnitine) recruits multiple substrate-binding proteins with strong specificity for distinct quaternary ammonium compounds. Mol. Microbiol. 2010, 75, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Beattie, G.A. Characterization of the osmoprotectant transporter OpuC from Pseudomonas syringae and demonstration that cystathionine-beta-synthase domains are required for its osmoregulatory function. J. Bacteriol. 2007, 189, 6901–6912. [Google Scholar] [CrossRef] [PubMed]
- Kleber, H.P. Bacterial carnitine metabolism. FEMS Microbiol. Lett. 1997, 147, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hanschmann, H.; Kleber, H.-P. Purification and characterization of d(+)-carnitine dehydrogenase from Agrobacterium sp.—A new enzyme of carnitine metabolism. Biochim. Biophys. Acta Protein Struct. Mol. Enzymol. 1997, 1337, 133–142. [Google Scholar] [CrossRef]
- Meadows, J.A.; Wargo, M.J. Characterization of Pseudomonas aeruginosa growth on O-acylcarnitines and identification of a short-chain acylcarnitine hydrolase. Appl. Environ. Microbiol. 2013, 79, 3355–3363. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.T.; Pocard, J.A.; Bernard, T.; Le Rudulier, D. Osmotic control of glycine betaine biosynthesis and degradation in Rhizobium meliloti. J. Bacteriol. 1988, 170, 3142–3149. [Google Scholar] [CrossRef] [PubMed]
- Seim, H.; Loster, H.; Claus, R.; Kleber, H.P.; Strack, E. Stimulation of the anaerobic growth of Salmonella typhimurium by reduction of l-carnitine, carnitine derivatives and structure-related trimethylammonium compounds. Arch. Microbiol. 1982, 132, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Seim, H.; Loster, H.; Kleber, H.P. Reductive metabolism of l-carnitine and structure-related trimethylammonium compounds in Escherichia coli. Acta Biol. Med. Ger. 1982, 41, 1009–1018. [Google Scholar] [PubMed]
- Aurich, H.; Kleber, H.P.; Schopp, W.D. An inducible carnitine dehydrogenase from Pseudomonas aeruginosa. Biochim. Biophys. Acta 1967, 139, 505–507. [Google Scholar] [CrossRef]
- Brown, A.D.; Simpson, J.R. Water relations of sugar-tolerant yeasts: The role of intracellular polyols. J. Gen. Microbiol. 1972, 72, 589–591. [Google Scholar] [CrossRef] [PubMed]
- Warren, C.R. High diversity of small organic N observed in soil water. Soil Biol. Biochem. 2013, 57 (Suppl. C), 444–450. [Google Scholar] [CrossRef]
- Warren, C.R. Quaternary ammonium compounds can be abundant in some soils and are taken up as intact molecules by plants. New Phytol. 2013, 198, 476–485. [Google Scholar] [CrossRef] [PubMed]
- Smiddy, M.; Sleator, R.D.; Patterson, M.F.; Hill, C.; Kelly, A.L. Role for compatible solutes glycine betaine and l-carnitine in Listerial barotolerance. Appl. Environ. Microbiol. 2004, 70, 7555–7557. [Google Scholar] [CrossRef] [PubMed]
- Kappes, R.M.; Bremer, E. Response of Bacillus subtilis to high osmolarity: Uptake of carnitine, crotonobetaine and γ-butyrobetaine via the ABC transport system OpuC. Microbiology 1998, 144, 83–90. [Google Scholar] [CrossRef]
- Angelidis, A.S.; Smith, G.M. Role of the glycine betaine and carnitine transporters in adaptation of Listeria monocytogenes to chill stress in defined medium. Appl. Environ. Microbiol. 2003, 69, 7492–7498. [Google Scholar] [CrossRef] [PubMed]
- Watson, D.; Sleator, R.D.; Casey, P.G.; Hill, C.; Gahan, C.G. Specific osmolyte transporters mediate bile tolerance in Listeria monocytogenes. Infect. Immun. 2009, 77, 4895–4904. [Google Scholar] [CrossRef] [PubMed]
- Gahan, C.G.; Hill, C. Listeria monocytogenes: Survival and adaptation in the gastrointestinal tract. Food Res. Int. 2014, 4, 9. [Google Scholar] [CrossRef] [PubMed]
- Wargo, M.J. Homeostasis and catabolism of choline and glycine betaine: Lessons from Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2013, 79, 2112–2120. [Google Scholar] [CrossRef] [PubMed]
- Malek, A.A.; Chen, C.; Wargo, M.J.; Beattie, G.A.; Hogan, D.A. Roles of three transporters, CbcXWV, BetT1, and BetT3, in Pseudomonas aeruginosa choline uptake for catabolism. J. Bacteriol. 2011, 193, 3033–3041. [Google Scholar] [CrossRef] [PubMed]
- Ziegler, C.; Bremer, E.; Kramer, R. The BCCT family of carriers: From physiology to crystal structure. Mol. Microbiol. 2010, 78, 13–34. [Google Scholar] [CrossRef] [PubMed]
- Williams, P.; Winzer, K.; Chan, W.C.; Camara, M. Look who’s talking: Communication and quorum sensing in the bacterial world. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2007, 362, 1119–1134. [Google Scholar] [CrossRef] [PubMed]
- Hamon, M.A.; Lazazzera, B.A. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol. Microbiol. 2001, 42, 1199–1209. [Google Scholar] [CrossRef] [PubMed]
- Auchtung, J.M.; Lee, C.A.; Grossman, A.D. Modulation of the ComA—Dependent quorum response in Bacillus subtilis by multiple Rap proteins and Phr peptides. J. Bacteriol. 2006, 188, 5273–5285. [Google Scholar] [CrossRef] [PubMed]
- Fujiya, M.; Musch, M.W.; Nakagawa, Y.; Hu, S.; Alverdy, J.; Kohgo, Y.; Schneewind, O.; Jabri, B.; Chang, E.B. The Bacillus subtilis quorum-sensing molecule CSF contributes to intestinal homeostasis via OCTN2, a host cell membrane transporter. Cell Host Microbe 2007, 1, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Lazazzera, B.A. The intracellular function of extracellular signaling peptides. Peptides 2001, 22, 1519–1527. [Google Scholar] [CrossRef]
- Tao, Y.; Drabik, K.A.; Waypa, T.S.; Musch, M.W.; Alverdy, J.C.; Schneewind, O.; Chang, E.B.; Petrof, E.O. Soluble factors from Lactobacillus GG activate MAPKs and induce cytoprotective heat shock proteins in intestinal epithelial cells. Am. J. Physiol. Cell Physiol. 2006, 290, C1018–C1030. [Google Scholar] [CrossRef] [PubMed]
- Williams, P. Bacillus subtilis: A shocking message from a probiotic. Cell Host Microbe 2007, 1, 248–249. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wan, Y.-J.Y. Nuclear receptors and inflammatory diseases. Exp. Biol. Med. 2008, 233, 496–506. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, G.; Petillo, O.; Margarucci, S.; Torpedine, A.; Calarco, A.; Koverech, A.; Boccia, A.; Paolella, G.; Peluso, G. Colon OCTN2 gene expression is up-regulated by peroxisome proliferator-activated receptor γ in humans and mice and contributes to local and systemic carnitine homeostasis. J. Biol. Chem. 2010, 285, 27078–27087. [Google Scholar] [CrossRef] [PubMed]
- Kalousek, F.; Darigo, M.D.; Rosenberg, L.E. Isolation and characterization of propionyl-CoA carboxylase from normal human liver. Evidence for a protomeric tetramer of nonidentical subunits. J. Biol. Chem. 1980, 255, 60–65. [Google Scholar] [PubMed]
- Chapman, K.A.; Gropman, A.; MacLeod, E.; Stagni, K.; Summar, M.L.; Ueda, K.; Mew, N.A.; Franks, J.; Island, E.; Matern, D.; et al. Acute Management of Propionic Acidemia. Mol. Genet. Metab. 2012, 105, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Roe, C.R.; Millington, D.S.; Maltby, D.A.; Bohan, T.P.; Hoppel, C.L. l-carnitine enhances excretion of propionyl coenzyme A as propionylcarnitine in propionic acidemia. J. Clin. Investig. 1984, 73, 1785–1788. [Google Scholar] [CrossRef] [PubMed]
- Higashimura, Y.; Naito, Y.; Takagi, T.; Uchiyama, K.; Mizushima, K.; Yoshikawa, T. Propionate promotes fatty acid oxidation through the up-regulation of Peroxisome proliferator-activated receptor alpha in intestinal epithelial cells. J. Nutr. Sci. Vitaminol. 2015, 61, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Kets, E.P.W.; de Bont, J.A.M. Effect of carnitines on Lactobacillus plantarum subjected to osmotic stress. FEMS Microbiol. Lett. 1997, 146, 205–209. [Google Scholar] [CrossRef]
- Borthakur, A.; Anbazhagan, A.N.; Kumar, A.; Raheja, G.; Singh, V.; Ramaswamy, K.; Dudeja, P.K. The probiotic Lactobacillus plantarum counteracts TNF-α-induced downregulation of SMCT1 expression and function. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, G928–G934. [Google Scholar] [CrossRef] [PubMed]
- Puddu, A.; Sanguineti, R.; Montecucco, F.; Viviani, G.L. Evidence for the gut microbiota short-chain fatty acids as key pathophysiological molecules improving diabetes. Mediat. Inflamm. 2014, 2014, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hadjiagapiou, C.; Schmidt, L.; Dudeja, P.K.; Layden, T.J.; Ramaswamy, K. Mechanism(s) of butyrate transport in Caco-2 cells: Role of monocarboxylate transporter 1. Am. J. Physiol. Gastrointest. Liver Physiol. 2000, 279, G775–G780. [Google Scholar] [CrossRef] [PubMed]
- Fritz, I.B. Carnitine and its role in fatty acid metabolism. Adv. Lipid Res. 1963, 1, 285–334. [Google Scholar] [PubMed]
- D’Argenio, G.; Calvani, M.; Casamassimi, A.; Petillo, O.; Margarucci, S.; Rienzo, M.; Peluso, I.; Calvani, R.; Ciccodicola, A.; Caporaso, N.; et al. Experimental colitis: Decreased Octn2 and Atb0+ expression in rat colonocytes induces carnitine depletion that is reversible by carnitine-loaded liposomes. FASEB J. 2006, 20, 2544–2546. [Google Scholar] [CrossRef] [PubMed]
- Zambell, K.L.; Fitch, M.D.; Fleming, S.E. Acetate and butyrate are the major substrates for de novo lipogenesis in rat colonic epithelial cells. J. Nutr. 2003, 133, 3509–3515. [Google Scholar] [CrossRef] [PubMed]
- Boren, J.; Lee, W.N.; Bassilian, S.; Centelles, J.J.; Lim, S.; Ahmed, S.; Boros, L.G.; Cascante, M. The stable isotope-based dynamic metabolic profile of butyrate-induced HT29 cell differentiation. J. Biol. Chem. 2003, 278, 28395–28402. [Google Scholar] [CrossRef] [PubMed]
- Peluso, G.; Petillo, O.; Margarucci, S.; Mingrone, G.; Greco, A.V.; Indiveri, C.; Palmieri, F.; Melone, M.A.; Reda, E.; Calvani, M. Decreased mitochondrial carnitine translocase in skeletal muscles impairs utilization of fatty acids in insulin-resistant patients. Front. Biosci. 2002, 7, a109–a116. [Google Scholar] [CrossRef] [PubMed]
- Peluso, G.; Nicolai, R.; Reda, E.; Benatti, P.; Barbarisi, A.; Calvani, M. Cancer and anticancer therapy-induced modifications on metabolism mediated by carnitine system. J. Cell Physiol. 2000, 182, 339–350. [Google Scholar] [CrossRef]
- McGarry, J.D.; Brown, N.F. The mitochondrial carnitine palmitoyltransferase system. From concept to molecular analysis. Eur. J. Biochem. 1997, 244, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, R.R.; Gandour, R.D.; van der Leij, F.R. Molecular enzymology of carnitine transfer and transport. Biochim. Biophys. Acta 2001, 1546, 21–43. [Google Scholar] [CrossRef]
- Stephens, F.B.; Constantin-Teodosiu, D.; Greenhaff, P.L. New insights concerning the role of carnitine in the regulation of fuel metabolism in skeletal muscle. J. Physiol. 2007, 581 Pt 2, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Jakobs, B.S.; Wanders, R.J. Fatty acid beta-oxidation in peroxisomes and mitochondria: The first, unequivocal evidence for the involvement of carnitine in shuttling propionyl-CoA from peroxisomes to mitochondria. Biochem. Biophys. Res. Commun. 1995, 213, 1035–1041. [Google Scholar] [CrossRef] [PubMed]
- Verhoeven, N.M.; Roe, D.S.; Kok, R.M.; Wanders, R.J.; Jakobs, C.; Roe, C.R. Phytanic acid and pristanic acid are oxidized by sequential peroxisomal and mitochondrial reactions in cultured fibroblasts. J. Lipid Res. 1998, 39, 66–74. [Google Scholar] [PubMed]
- Carter, A.L.; Abney, T.O.; Lapp, D.F. Biosynthesis and metabolism of carnitine. J. Child Neurol. 1995, 10, S3–S7. [Google Scholar] [CrossRef] [PubMed]
- Duran, M.; Loof, N.E.; Ketting, D.; Dorland, L. Secondary carnitine deficiency. J. Clin. Chem. Clin. Biochem. 1990, 28, 359–363. [Google Scholar] [PubMed]
- Hulse, J.D.; Ellis, S.R.; Henderson, L.M. Carnitine biosynthesis. beta-Hydroxylation of trimethyllysine by an alpha-ketoglutarate-dependent mitochondrial dioxygenase. J. Biol. Chem. 1978, 253, 1654–1659. [Google Scholar] [PubMed]
- Sachan, D.S.; Broquist, H.P. Synthesis of carnitine from ε-N-trimethyllysine in post mitochondrial fractions of Neurospora crassa. Biochem. Biophys. Res. Commun. 1980, 96, 870–875. [Google Scholar] [CrossRef]
- Sachan, D.S.; Hoppel, C.L. Carnitine biosynthesis. Hydroxylation of N6-trimethyl-lysine to 3-hydroxy-N6-trimethyl-lysine. Biochem. J. 1980, 188, 529–534. [Google Scholar] [CrossRef] [PubMed]
- Stein, R.; Englard, S. Properties of rat 6-N-trimethyl-l-lysine hydroxylases: Similarities among the kidney, liver, heart, and skeletal muscle activities. Arch. Biochem. Biophys. 1982, 217, 324–331. [Google Scholar] [CrossRef]
- Torre, M.; Rodriguez, A.R.; Saura-Calixto, F. Interactions of Fe(II), Ca(II) and Fe(III) with high dietary fibre materials: A physicochemical approach. Food Chem. 1995, 54, 23–31. [Google Scholar] [CrossRef]
- Baye, K.; Guyot, J.P.; Mouquet-Rivier, C. The unresolved role of dietary fibers on mineral absorption. Crit. Rev. Food Sci. Nutr. 2017, 57, 949–957. [Google Scholar] [CrossRef] [PubMed]
- Van der Klis, J.D.; Kwakernaak, C.; de Wit, W. Effects of endoxylanase addition to wheat-based diets on physico-chemical chyme conditions and mineral absorption in broilers. Anim. Feed Sci. Technol. 1995, 51, 15–27. [Google Scholar] [CrossRef]
- Guillon, F.; Champ, M. Structural and physical properties of dietary fibres, and consequences of processing on human physiology. Food Res. Int. 2000, 33, 233–245. [Google Scholar] [CrossRef]
- Debon, S.J.J.; Tester, R.F. In vitro binding of calcium, iron and zinc by non-starch polysaccharides. Food Chem. 2001, 73, 401–410. [Google Scholar] [CrossRef]
- Miyada, T.; Nakajima, A.; Ebihara, K. Iron bound to pectin is utilised by rats. Br. J. Nutr. 2011, 106, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Atallah, M.T.; Amarasiriwardena, C.; Barnes, R. Pectin with low molecular weight and high degree of esterification increases absorption of 58Fe in growing rats. J. Nutr. 1996, 126, 1883–1890. [Google Scholar] [PubMed]
- Liu, J.; Liu, X.; Xiong, X.Q.; Yang, T.; Cui, T.; Hou, N.L.; Lai, X.; Liu, S.; Guo, M.; Liang, X.H.; et al. Effect of vitamin A supplementation on gut microbiota in children with autism spectrum disorders—A pilot study. BMC Microbiol. 2017, 17, 204. [Google Scholar] [CrossRef] [PubMed]
- Skrypnik, K.; Suliburska, J. Association between the gut microbiota and mineral metabolism. J. Sci. Food Agric. 2017. [Google Scholar] [CrossRef] [PubMed]
- Clouatre, D.; Bell, S. Is l-carnitine the link between red meat and heart disease? J. Nutr. Food Sci. 2013, 3, e119. [Google Scholar] [CrossRef]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576–585. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghonimy, A.; Zhang, D.M.; Farouk, M.H.; Wang, Q. The Impact of Carnitine on Dietary Fiber and Gut Bacteria Metabolism and Their Mutual Interaction in Monogastrics. Int. J. Mol. Sci. 2018, 19, 1008. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19041008
Ghonimy A, Zhang DM, Farouk MH, Wang Q. The Impact of Carnitine on Dietary Fiber and Gut Bacteria Metabolism and Their Mutual Interaction in Monogastrics. International Journal of Molecular Sciences. 2018; 19(4):1008. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19041008
Chicago/Turabian StyleGhonimy, Abdallah, Dong Ming Zhang, Mohammed Hamdy Farouk, and Qiuju Wang. 2018. "The Impact of Carnitine on Dietary Fiber and Gut Bacteria Metabolism and Their Mutual Interaction in Monogastrics" International Journal of Molecular Sciences 19, no. 4: 1008. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19041008