Imine Deaminase Activity and Conformational Stability of UK114, the Mammalian Member of the Rid Protein Family Active in Amino Acid Metabolism
Abstract
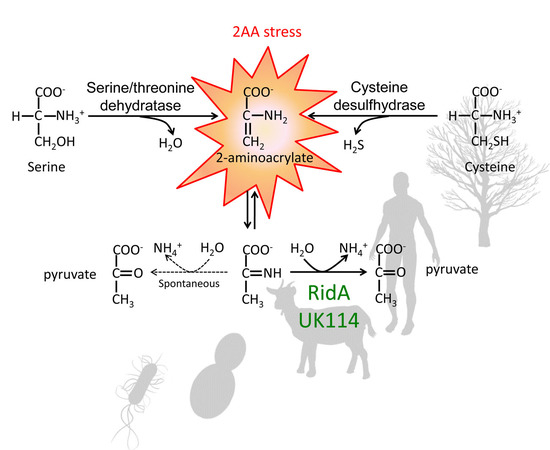
:1. Introduction
2. Results
2.1. Characterization of UK114 from Capra hircus (Goat)
2.2. Expression and Purification of a Recombinant Goat UK114
2.3. C. hircus UK114 Is a Deiminase
Specificity of UK114 toward Different Imino Acids
2.4. Study of the Conformational Stability of Goat UK114
2.5. Behavior of the Conformational Stability and Enzymatic Activity of UK114 Upon Long-Term Incubation at High Temperature
3. Discussion
4. Materials and Methods
4.1. Construct for the Expression of Goat UK114 in E. coli
4.2. Protein Production and Purification
4.3. Analytical Gel Filtration
4.4. Protein Purity Determination by Nanoscale Liquid Chromatography—Mass Spectrometry (LC-MS)
4.5. Protein Conformational Integrity by Nanoscale Electrospray Mass Spectrometry (NSI-MS)
4.6. Protein Sequencing by Nanoscale Liquid Chromatography Tandem Mass Spectrometry (LC-NSI-MS/MS)
4.7. Activity Assays of UK114 Deiminase Activity
4.8. Circular Dichroism
4.9. Heat Inactivation of UK114 and Data Analysis
4.10. Treatments of UK114 with Perchloric Acid and with a Freeze-Drying-Reconstitutionprocess
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| Rid | Reactive intermediate deaminase |
| PLP | Pyridoxal 5′-phosphate |
| 2AA | 2-aminoacrylate |
| MS | Mass spectrometry |
| CD | Circular dichroism |
| LAAO | l-amino acid oxidase |
| DAAO | d-amino acid oxidase |
References
- Lambrecht, J.A.; Flynn, J.M.; Downs, D.M. Conserved YjgF protein family deaminates reactive enamine/imine intermediates of pyridoxal 5’-phosphate (PLP)-dependent enzyme reactions. J. Biol. Chem. 2012, 287, 3454–3461. [Google Scholar] [CrossRef] [PubMed]
- Lambrecht, J.A.; Browne, B.A.; Downs, D.M. Members of the YjgF/YER057c/UK114 family of proteins inhibit phosphoribosylamine synthesis in vitro. J. Biol. Chem. 2010, 285, 34401–34407. [Google Scholar] [CrossRef] [PubMed]
- Lambrecht, J.A.; Schmitz, G.E.; Downs, D.M. RidA proteins prevent metabolic damage inflicted by PLP-dependent dehydratases in all domains of life. mBio 2013, 4, e00033-13. [Google Scholar] [CrossRef] [PubMed]
- Downs, D.M.; Ernst, D.C. From microbiology to cancer biology: The Rid protein family prevents cellular damage caused by endogenously generated reactive nitrogen species. Mol. Microbiol. 2015, 96, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Ernst, D.C.; Downs, D.M. 2-Aminoacrylate Stress Induces a Context-Dependent Glycine Requirement in RidA Strains of Salmonella enterica. J. Bacteriol. 2015, 198, 536–543. [Google Scholar] [CrossRef] [PubMed]
- Niehaus, T.D.; Gerdes, S.; Hodge-Hanson, K.; Zhukov, A.; Cooper, A.J.; ElBadawi-Sidhu, M.; Fiehn, O.; Downs, D.M.; Hanson, A.D. Genomic and experimental evidence for multiple metabolic functions in the RidA/YjgF/YER057c/UK114 (Rid) protein family. BMC Genom. 2015, 16, 382. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.M.; Downs, D.M. In the absence of RidA, endogenous 2-aminoacrylate inactivates alanine racemases by modifying the pyridoxal 5′-phosphate cofactor. J. Bacteriol. 2013, 195, 3603–3609. [Google Scholar] [CrossRef] [PubMed]
- Cavallini, D.; Federici, G.; Bossa, F.; Granata, F. The protective effect of thiosulfate upon the inactivation of aspartate aminotransferase by aminoacrylic-acid-producing substrates. Eur. J. Biochem. 1973, 39, 301–304. [Google Scholar] [CrossRef] [PubMed]
- Ernst, D.C.; Anderson, M.E.; Downs, D.M. l-2,3-diaminopropionate generates diverse metabolic stresses in Salmonella enterica. Mol. Microbiol. 2016, 101, 210–223. [Google Scholar] [CrossRef] [PubMed]
- Danson, J.W.; Trawick, M.L.; Cooper, A.J. Spectrophotometric assays for l-lysine alpha-oxidase and gamma-glutamylamine cyclotransferase. Anal. Biochem. 2002, 303, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Hafner, E.W.; Wellner, D. Reactivity of the imino acids formed in the amino acid oxidase reaction. Biochemistry 1979, 18, 411–417. [Google Scholar] [CrossRef] [PubMed]
- Datta, P.; Bhadra, R. Biodegradative threonine dehydratase. Reduction of ferricyanide by an intermediate of the enzyme-catalyzed reaction. Eur. J. Biochem. 1978, 91, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Hillebrand, G.G.; Dye, J.L.; Suelter, C.H. Formation of an intermediate and its rate of conversion to pyruvate during the tryptophanase-catalyzed degradation of S-o-nitrophenyl-l-cysteine. Biochemistry 1979, 18, 1751–1755. [Google Scholar] [CrossRef] [PubMed]
- Borchert, A.J.; Downs, D.M. Endogenously generated 2-aminoacrylate inhibits motility in Salmonella enterica. Sci. Rep. 2017, 7, 12971. [Google Scholar] [CrossRef] [PubMed]
- Hodge-Hanson, K.M.; Downs, D.M. Members of the Rid protein family have broad imine deaminase activity and can accelerate the Pseudomonas aeruginosa d-arginine dehydrogenase (DauA) reaction in vitro. PLoS ONE 2017, 12, e0185544. [Google Scholar] [CrossRef] [PubMed]
- Borchert, A.J.; Downs, D.M. The Response to 2-Aminoacrylate Differs in Escherichia coli and Salmonella enterica, despite Shared Metabolic Components. J. Bacteriol. 2017, 199. [Google Scholar] [CrossRef] [PubMed]
- Niehaus, T.D.; Nguyen, T.N.; Gidda, S.K.; ElBadawi-Sidhu, M.; Lambrecht, J.A.; McCarty, D.R.; Downs, D.M.; Cooper, A.J.; Fiehn, O.; Mullen, R.T.; et al. Arabidopsis and maize RidA proteins preempt reactive enamine/imine damage to branched-chain amino acid biosynthesis in plastids. Plant Cell 2014, 26, 3010–3022. [Google Scholar] [CrossRef] [PubMed]
- Manjasetty, B.A.; Delbruck, H.; Pham, D.T.; Mueller, U.; Fieber-Erdmann, M.; Scheich, C.; Sievert, V.; Bussow, K.; Niesen, F.H.; Weihofen, W.; et al. Crystal structure of Homo sapiens protein hp14.5. Proteins 2004, 54, 797–800. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zeng, J.; Chen, X.; Xie, W. Crystal structures of RidA, an important enzyme for the prevention of toxic side products. Sci. Rep. 2016, 6, 30494. [Google Scholar] [CrossRef] [PubMed]
- Burman, J.D.; Stevenson, C.E.; Sawers, R.G.; Lawson, D.M. The crystal structure of Escherichia coli TdcF, a member of the highly conserved YjgF/YER057c/UK114 family. BMC Struct. Biol. 2007, 7, 30. [Google Scholar] [CrossRef] [PubMed]
- Colombo, I.; Ceciliani, F.; Ronchi, S.; Bartorelli, A.; Berra, B. cDNA cloning and Escherichia coli expression of UK114 tumor antigen. Biochim. Biophys. Acta 1998, 1442, 49–59. [Google Scholar] [CrossRef]
- Panerai, A.E.; Sacerdote, P.; Bianchi, M.; Nicoletti, F.; Manfredi, B.; Gaspani, L.; Bartorelli, A.; Ceciliani, F.; Ronchi, S. Chronic administration of UK-114, a multifunctional emerging protein, modulates the Th1/Th2 cytokine pattern and experimental autoimmune diseases. Ann. N. Y. Acad. Sci. 1999, 876, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Funaro, A.; Horenstein, A.L.; Ghisolfi, G.; Bussolati, B.; Bartorelli, A.; Bussolati, G. Identification of a 220-kDa membrane tumor-associated antigen by human anti-UK114 monoclonal antibodies selected from the immunoglobulin repertoire of a cancer patient. Exp. Cell Res. 1999, 247, 441–450. [Google Scholar] [CrossRef] [PubMed]
- Bartorelli, A.; Berra, B.; Ronchi, S.; Biancardi, C.; Cavalca, V.; Ballo, M.; Ferrara, R.; Botta, M.; Arzani, C.; Clemente, C. Immunocytochemical reactivity of mammalian liver antigen (UK101) in human tumors and non neoplastic tissues. J. Tumor Marker Oncol. 1994, 9, 37–48. [Google Scholar]
- Bartorelli, A.; Bussolati, B.; Millesimo, M.; Gugliotta, P.; Bussolati, G. Antibody-dependent cytotoxic activity on human cancer cells expressing UK 114 tumor membrane antigen. Int. J. Oncol. 1996, 8, 543–548. [Google Scholar] [CrossRef] [PubMed]
- Bartorelli, A.; Biancardi, C.; Cavalca, V.; Ferrara, R.; Botta, M.; Arzani, C.; Colombo, I.; Berra, B.; Ceciliani, F.; Ronchi, S.; et al. Purification and partial characterization of proteins present in a perchloric acid extract of goat liver (UK101). J. Tumor Marker Oncol. 1996, 11, 57–61. [Google Scholar]
- Ceciliani, F.; Biancardi, C.; Cavalca, V.; Ferrara, R.; Botta, M.; Bailo, M.; Arzani, C.; Berra, B.; Ronchi, S.; Bartorelli, A. Structural characterization of the small molecular weight proteins present in UK101. J. Tumor Marker Oncol. 1996, 11, 63–66. [Google Scholar]
- Bartorelli, A.; Turiano, A. Substances of Polypeptide Nature Useful in Human Therapy. Patent WO1992010197A1, 12 November 1990. [Google Scholar]
- Ceciliani, F.; Faotto, L.; Negri, A.; Colombo, I.; Berra, B.; Bartorelli, A.; Ronchi, S. The primary structure of UK114 tumor antigen. FEBS Lett. 1996, 393, 147–150. [Google Scholar] [CrossRef]
- Bussolati, G.; Geuna, M.; Bussolati, B.; Millesimo, M.; Botta, M.; Bartorelli, A. Cytolytic and tumor inhibitory antibodies against UK114 protein in the sera of cancer patients. Int. J. Oncol. 1997, 10, 779–785. [Google Scholar] [CrossRef] [PubMed]
- Schmiedeknecht, G.; Kerkhoff, C.; Orso, E.; Stohr, J.; Aslanidis, C.; Nagy, G.M.; Knuechel, R.; Schmitz, G. Isolation and characterization of a 14.5-kDa trichloroacetic-acid-soluble translational inhibitor protein from human monocytes that is upregulated upon cellular differentiation. Eur. J. Biochem. 1996, 242, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Pozdniakovaite, N.; Popendikyte, V. DNA methylation differences in human p14.5 gene promoter region in normal and proliferating cells. Dev. Growth Differ. 2005, 47, 493–499. [Google Scholar] [CrossRef] [PubMed]
- Oxelmark, E.; Marchini, A.; Malanchi, I.; Magherini, F.; Jaquet, L.; Hajibagheri, M.A.; Blight, K.J.; Jauniaux, J.C.; Tommasino, M. Mmf1p, a novel yeast mitochondrial protein conserved throughout evolution and involved in maintenance of the mitochondrial genome. Mol. Cell. Biol. 2000, 20, 7784–7797. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; Yoshikawa, H.; Shirahige, K. A member of the YER057c/YjgF/Uk114 family links isoleucine biosynthesis and intact mitochondria maintenance in Saccharomyces cerevisiae. Genes Cells Devoted Mol. Cell. Mech. 2001, 6, 507–517. [Google Scholar] [CrossRef]
- Ernst, D.C.; Downs, D.M. Mmf1p Couples Amino Acid Metabolism to Mitochondrial DNA Maintenance in Saccharomyces cerevisiae. mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Antonenkov, V.D.; Ohlmeier, S.; Sormunen, R.T.; Hiltunen, J.K. UK114, a YjgF/Yer057p/UK114 family protein highly conserved from bacteria to mammals, is localized in rat liver peroxisomes. Biochem. Biophys. Res. Commun. 2007, 357, 252–257. [Google Scholar] [CrossRef] [PubMed]
- ElRamlawy, K.G.; Fujimura, T.; Baba, K.; Kim, J.W.; Kawamoto, C.; Isobe, T.; Abe, T.; Hodge-Hanson, K.; Downs, D.M.; Refaat, I.H.; et al. Der f 34, a Novel Major House Dust Mite Allergen Belonging to a Highly Conserved Rid/YjgF/YER057c/UK114 Family of Imine Deaminases. J. Biol. Chem. 2016, 291, 21607–21615. [Google Scholar] [CrossRef] [PubMed]
- Morishita, R.; Kawagoshi, A.; Sawasaki, T.; Madin, K.; Ogasawara, T.; Oka, T.; Endo, Y. Ribonuclease activity of rat liver perchloric acid-soluble protein, a potent inhibitor of protein synthesis. J. Biol. Chem. 1999, 274, 20688–20692. [Google Scholar] [CrossRef] [PubMed]
- Oka, T.; Tsuji, H.; Noda, C.; Sakai, K.; Hong, Y.M.; Suzuki, I.; Munoz, S.; Natori, Y. Isolation and characterization of a novel perchloric acid-soluble protein inhibiting cell-free protein synthesis. J. Biol. Chem. 1995, 270, 30060–30067. [Google Scholar] [PubMed]
- Samuel, S.J.; Tzung, S.P.; Cohen, S.A. Hrp12, a novel heat-responsive, tissue-specific, phosphorylated protein isolated from mouse liver. Hepatology 1997, 25, 1213–1222. [Google Scholar] [CrossRef] [PubMed]
- Park, O.H.; Park, J.; Yu, M.; An, H.T.; Ko, J.; Kim, Y.K. Identification and molecular characterization of cellular factors required for glucocorticoid receptor-mediated mRNA decay. Genes Dev. 2016, 30, 2093–2105. [Google Scholar] [CrossRef] [PubMed]
- Deriu, D.; Briand, C.; Mistiniene, E.; Naktinis, V.; Grutter, M.G. Structure and oligomeric state of the mammalian tumour-associated antigen UK114. Acta Crystallogr. Sect. D Biol. Crystallogr. 2003, 59, 1676–1678. [Google Scholar] [CrossRef]
- Mistiniene, E.; Pozdniakovaite, N.; Popendikyte, V.; Naktinis, V. Structure-based ligand binding sites of protein p14.5, a member of protein family YER057c/YIL051c/YjgF. Int. J. Biol. Macromol. 2005, 37, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Mistiniene, E.; Luksa, V.; Sereikaite, J.; Naktinis, V. Oligomeric assembly and ligand binding of the members of protein family YER057c/YIL051c/YJGF. Bioconjug. Chem. 2003, 14, 1243–1252. [Google Scholar] [CrossRef] [PubMed]
- Degani, G.; Popolo, L.; Department of Biosciences, University of Milan, Milan, Italy. Thermofluor analysis of goat UK114. Unpublished work. 2017. [Google Scholar]
- Barbiroli, A.; Department of Food, Environmental and Nutritional Sciences, University of Milan, Milan, Italy. Circular dichroism studies of the folding, thermal unfolding and refolding properties of freshly purified or freeze-dried UK114. Unpublished work. 2017. [Google Scholar]
- Barbiroli, A.; Popolo, L.; Vanoni, M.A. Analysis of the conformational stability and activity of UK114 after perchloric acid treatment. Unpublished work. 2017. [Google Scholar]
- Mehta, P.K.; Christen, P. The molecular evolution of pyridoxal-5’-phosphate-dependent enzymes. Adv. Enzymol. Relat. Areas Mol. Biol. 2000, 74, 129–184. [Google Scholar] [PubMed]
- Di Salvo, M.L.; Budisa, N.; Contestabile, R. PLP-Dependent Enzymes: A Powerful Tool for Metabolic Synthesis of non-Canonical Amino Acids. In Proceedings of the Beilsten Bozen Synposium on Molecular Engineering and Control, Prien (Chiemsee), Germany, 14–18 May 2012. [Google Scholar]
- Finney, J.; Moon, H.J.; Ronnebaum, T.; Lantz, M.; Mure, M. Human copper-dependent amine oxidases. Arch. Biochem. Biophys. 2014, 546, 19–32. [Google Scholar] [CrossRef] [PubMed]
- Binda, C.; Milczek, E.M.; Bonivento, D.; Wang, J.; Mattevi, A.; Edmondson, D.E. Lights and shadows on monoamine oxidase inhibition in neuroprotective pharmacological therapies. Curr. Top. Med. Chem. 2011, 11, 2788–2796. [Google Scholar] [CrossRef] [PubMed]
- Paul, P.; de Belleroche, J. The role of d-amino acids in amyotrophic lateral sclerosis pathogenesis: A review. Amino Acids 2012, 43, 1823–1831. [Google Scholar] [CrossRef] [PubMed]
- Castellano, F.; Molinier-Frenkel, V. An Overview of l-Amino Acid Oxidase Functions from Bacteria to Mammals: Focus on the Immunoregulatory Phenylalanine Oxidase IL4I1. Molecules 2017, 22, 2151. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, P.F. Oxidation of amines by flavoproteins. Arch. Biochem. Biophys. 2010, 493, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Edmondson, D.E. Hydrogen peroxide produced by mitochondrial monoamine oxidase catalysis: Biological implications. Curr. Pharm. Des. 2014, 20, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.L.; Grant, G.A. Mutagenic and chemical analyses provide new insight into enzyme activation and mechanism of the type 2 iron-sulfur l-serine dehydratase from Legionella pneumophila. Arch. Biochem. Biophys. 2016, 596, 108–117. [Google Scholar] [CrossRef] [PubMed]
- Pollegioni, L.; Molla, G. New biotech applications from evolved d-amino acid oxidases. Trends Biotechnol. 2011, 29, 276–283. [Google Scholar] [CrossRef] [PubMed]
- Keller, B.O.; Sui, J.; Young, A.B.; Whittal, R.M. Interferences and contaminants encountered in modern mass spectrometry. Anal. Chim. Acta 2008, 627, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Leney, A.C.; Heck, A.J. Native Mass Spectrometry: What is in the Name? J. Am. Soc. Mass Spectrom. 2017, 28, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Regazzoni, L.; Colombo, S.; Mazzolari, A.; Vistoli, G.; Carini, M. Serum albumin as a probe for testing the selectivity of irreversible cysteine protease inhibitors: The case of vinyl sulfones. J. Pharm. Biomed. Anal. 2016, 124, 294–302. [Google Scholar] [CrossRef] [PubMed]
- Vanoni, M.A.; Cosma, A.; Mazzeo, D.; Mattevi, A.; Todone, F.; Curti, B. Limited proteolysis and X-ray crystallography reveal the origin of substrate specificity and of the rate-limiting product release during oxidation of d-amino acids catalyzed by mammalian d-amino acid oxidase. Biochemistry 1997, 36, 5624–5632. [Google Scholar] [CrossRef] [PubMed]
- Curti, B.; Ronchi, S.; Branzoli, U.; Ferri, G.; Williams, C.H., Jr. Improved purification, amino acid analysis and molecular weight of homogenous d-amino acid oxidase from pig kidney. Biochim. Biophys. Acta 1973, 327, 266–273. [Google Scholar] [CrossRef]
- Kelly, S.M.; Jess, T.J.; Price, N.C. How to study proteins by circular dichroism. Biochim. Biophys. Acta 2005, 1751, 119–139. [Google Scholar] [CrossRef] [PubMed]
- Bohm, G.; Muhr, R.; Jaenicke, R. Quantitative analysis of protein far UV circular dichroism spectra by neural networks. Protein Eng. 1992, 5, 191–195. [Google Scholar] [CrossRef] [PubMed]







| Amino Acid | K50, μM | 100/K50, μM−1 |
|---|---|---|
| Ala | 0.29 ± 0.02 | 344 ± 24 |
| Leu | 0.58 ± 0.03 | 172 ± 9 |
| l-Met | 0.41 ± 0.03 | 208 ± 17 |
| l-Gln | 2.56 ± 0.11 | 39 ± 2 |
| l-Phe | ND | 2.5 ± 0.2 |
| l-His | ND | ND |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Degani, G.; Barbiroli, A.; Regazzoni, L.; Popolo, L.; Vanoni, M.A. Imine Deaminase Activity and Conformational Stability of UK114, the Mammalian Member of the Rid Protein Family Active in Amino Acid Metabolism. Int. J. Mol. Sci. 2018, 19, 945. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19040945
Degani G, Barbiroli A, Regazzoni L, Popolo L, Vanoni MA. Imine Deaminase Activity and Conformational Stability of UK114, the Mammalian Member of the Rid Protein Family Active in Amino Acid Metabolism. International Journal of Molecular Sciences. 2018; 19(4):945. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19040945
Chicago/Turabian StyleDegani, Genny, Alberto Barbiroli, Luca Regazzoni, Laura Popolo, and Maria Antonietta Vanoni. 2018. "Imine Deaminase Activity and Conformational Stability of UK114, the Mammalian Member of the Rid Protein Family Active in Amino Acid Metabolism" International Journal of Molecular Sciences 19, no. 4: 945. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19040945