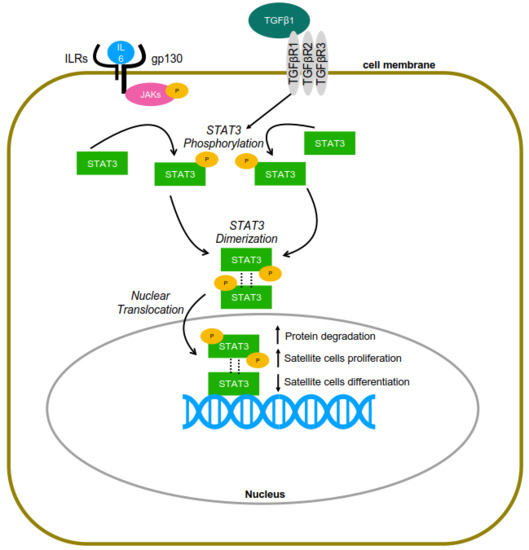
STAT3 in Skeletal Muscle Function and Disorders
Abstract
:1. Introduction
2. STAT3 Modulates Satellite Cell Myogenic Capacity and Self-Renewal
3. STAT3 Signaling in Response to Physiological Stimuli
4. STAT3 Signaling Contributes to Muscle Wasting in Cancer Cachexia
5. STAT3 Signaling in Inflammatory Myopathies
6. STAT3 Activation in Muscular Dystrophies
7. STAT3 and TGF-β1 Interplay in Muscle and Other Cell Types
8. Evidence of STAT3 Activation by TGF-β1 in Skeletal Muscle
9. Summary
Acknowledgments
Conflicts of Interest
References
- Schiaffino, S.; Dyar, K.A.; Ciciliot, S.; Blaauw, B.; Sandri, M. Mechanisms Regulating Skeletal Muscle Growth and Atrophy. FEBS J. 2013, 280, 4294–4314. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. Regulation of Mtorc1 and Its Impact on Gene Expression at a Glance. J. Cell Sci. 2013, 126, 1713–1719. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.; McLeod, L.; Alhayyani, S.; Szczepny, A.; Watkins, D.N.; Chen, W.; Enriori, P.; Ferlin, W.; Ruwanpura, S.; Jenkins, B.J. Blockade of the IL-6 Trans-Signalling/STAT3 Axis Suppresses Cachexia in Kras-Induced Lung Adenocarcinoma. Oncogene 2017, 36, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Zimmers, T.A.; Fishel, M.L.; Bonetto, A. STAT3 in the Systemic Inflammation of Cancer Cachexia. Semin. Cell Dev. Biol. 2016, 54, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Guadagnin, E.; Narola, J.; Bonnemann, C.G.; Chen, Y.W. Tyrosine 705 Phosphorylation of STAT3 Is Associated with Phenotype Severity in TGFbeta1 Transgenic Mice. BioMed Res. Int. 2015, 2015, 843743. [Google Scholar] [CrossRef] [PubMed]
- Akira, S.; Nishio, Y.; Inoue, M.; Wang, X.J.; Wei, S.; Matsusaka, T.; Yoshida, K.; Sudo, T.; Naruto, M.; Kishimoto, T. Molecular Cloning of APRF, a Novel IFN-Stimulated Gene Factor 3 P91-Related Transcription Factor Involved in the Gp130-Mediated Signaling Pathway. Cell 1994, 77, 63–71. [Google Scholar] [CrossRef]
- Nunes, A.M.; Wuebbles, R.D.; Sarathy, A.; Fontelonga, T.M.; Deries, M.; Burkin, D.J.; Thorsteinsdottir, S. Impaired Fetal Muscle Development and JAK-STAT Activation Mark Disease Onset and Progression in a Mouse Model for Merosin-Deficient Congenital Muscular Dystrophy. Hum. Mol. Genet. 2017, 26, 2018–2033. [Google Scholar] [CrossRef] [PubMed]
- Wada, E.; Tanihata, J.; Iwamura, A.; Takeda, S.; Hayashi, Y.K.; Matsuda, R. Treatment with the Anti-IL-6 Receptor Antibody Attenuates Muscular Dystrophy via Promoting Skeletal Muscle Regeneration in Dystrophin-/Utrophin-Deficient Mice. Skelet. Muscle 2017, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- Liang, F.; Li, T.; Azuelos, I.; Giordano, C.; Liang, H.; Hussain, S.N.; Matecki, S.; Petrof, B.J. Ventilator-Induced Diaphragmatic Dysfunction in Mdx Mice. Muscle Nerve 2018, 57, 442–448. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, Y.; Chen, J.; Hu, C.; Teng, M.; Jiao, K.; Shen, Z.; Zhu, D.; Yue, J.; Li, Z.; et al. The ROS-Mediated Activation of IL-6/STAT3 Signaling Pathway Is Involved in the 27-Hydroxycholesterol-Induced Cellular Senescence in Nerve Cells. Toxicol. In Vitro 2017, 45, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Benito, C.; Davis, C.M.; Gomez-Sanchez, J.A.; Turmaine, M.; Meijer, D.; Poli, V.; Mirsky, R.; Jessen, K.R. STAT3 Controls the Long-Term Survival and Phenotype of Repair Schwann Cells during Nerve Regeneration. J. Neurosci. 2017, 37, 4255–4269. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.H.; Xiang, Y.; Li, H.; Zheng, L.; Xu, Y.; Yu, C.X.; Li, J.P.; Zhang, X.Y.; Xing, W.B.; Cao, D.S.; et al. VEGF-a Stimulates STAT3 Activity via Nitrosylation of Myocardin to Regulate the Expression of Vascular Smooth Muscle Cell Differentiation Markers. Sci. Rep. 2017, 7, 2660. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Lin, A.; Jiang, N.; Yan, H.; Ni, Z.; Qian, J.; Fang, W. Interleukin-6 Trans-Signalling Induces Vascular Endothelial Growth Factor Synthesis Partly via Janus Kinases-STAT3 Pathway in Human Mesothelial Cells. Nephrology (Carlton) 2017, 22, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Mauro, A. Satellite Cell of Skeletal Muscle Fibers. J. Biophys. Biochem. Cytol. 1961, 9, 493–495. [Google Scholar] [CrossRef] [PubMed]
- Moss, F.P.; Leblond, C.P. Nature of Dividing Nuclei in Skeletal Muscle of Growing Rats. J. Cell Biol. 1970, 44, 459–462. [Google Scholar] [CrossRef] [PubMed]
- Schultz, E.; Jaryszak, D.L.; Valliere, C.R. Response of Satellite Cells to Focal Skeletal Muscle Injury. Muscle Nerve 1985, 8, 217–222. [Google Scholar] [CrossRef] [PubMed]
- Snow, M.H. Myogenic Cell Formation in Regenerating Rat Skeletal Muscle Injured by Mincing. II. An Autoradiographic Study. Anat. Rec. 1977, 188, 201–217. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Price, F.; Rudnicki, M.A. Satellite Cells and the Muscle Stem Cell Niche. Physiol. Rev. 2013, 93, 23–67. [Google Scholar] [CrossRef] [PubMed]
- Tierney, M.T.; Aydogdu, T.; Sala, D.; Malecova, B.; Gatto, S.; Puri, P.L.; Latella, L.; Sacco, A. STAT3 Signaling Controls Satellite Cell Expansion and Skeletal Muscle Repair. Nat. Med. 2014, 20, 1182–1186. [Google Scholar] [CrossRef] [PubMed]
- Price, F.D.; von Maltzahn, J.; Bentzinger, C.F.; Dumont, N.A.; Yin, H.; Chang, N.C.; Wilson, D.H.; Frenette, J.; Rudnicki, M.A. Inhibition of JAK-STAT Signaling Stimulates Adult Satellite Cell Function. Nat. Med. 2014, 20, 1174–1181. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Xiao, F.; Wang, G.; Wei, X.; Jiang, L.; Chen, Y.; Zhu, L.; Wang, H.; Diao, Y.; Wang, H.; et al. STAT3 Regulates Self-Renewal of Adult Muscle Satellite Cells During Injury-Induced Muscle Regeneration. Cell Rep. 2016, 16, 2102–2115. [Google Scholar] [CrossRef] [PubMed]
- Begue, G.; Douillard, A.; Galbes, O.; Rossano, B.; Vernus, B.; Candau, R.; Py, G. Early Activation of Rat Skeletal Muscle IL-6/STAT1/STAT3 Dependent Gene Expression in Resistance Exercise Linked to Hypertrophy. PLoS ONE 2013, 8, e57141. [Google Scholar] [CrossRef] [PubMed]
- Trenerry, M.K.; Carey, K.A.; Ward, A.C.; Cameron-Smith, D. STAT3 Signaling Is Activated in Human Skeletal Muscle Following Acute Resistance Exercise. J. Appl. Physiol. (1985) 2007, 102, 1483–1489. [Google Scholar] [CrossRef] [PubMed]
- Toth, K.G.; McKay, B.R.; de Lisio, M.; Little, J.P.; Tarnopolsky, M.A.; Parise, G. IL-6 Induced STAT3 Signalling Is Associated with the Proliferation of Human Muscle Satellite Cells Following Acute Muscle Damage. PLoS ONE 2011, 6, e17392. [Google Scholar] [CrossRef] [PubMed]
- Glund, S.; Deshmukh, A.; Long, Y.C.; Moller, T.; Koistinen, H.A.; Caidahl, K.; Zierath, J.R.; Krook, A. Interleukin-6 Directly Increases Glucose Metabolism in Resting Human Skeletal Muscle. Diabetes 2007, 56, 1630–1637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Schindler, J.; Esparza, M.C.; McKendry, J.; Breen, L.; Philp, A.; Schenk, S. Overload-Mediated Skeletal Muscle Hypertrophy Is Not Impaired by Loss of Myofiber STAT3. Am. J. Physiol. Cell Physiol. 2017, 313, C257–C261. [Google Scholar] [CrossRef] [PubMed]
- Amorese, A.J.; Spangenburg, E.E. Defining the Status Quo in Muscle Hypertrophy. Focus on “Overload-Mediated Skeletal Muscle Hypertrophy Is Not Impaired by Loss of Myofiber STAT3”. Am. J. Physiol. Cell Physiol. 2017, 313, C255–C256. [Google Scholar] [CrossRef] [PubMed]
- Van de Vyver, M.; Engelbrecht, L.; Smith, C.; Myburgh, K.H. Neutrophil and Monocyte Responses to Downhill Running: Intracellular Contents of MPO, IL-6, IL-10, pstat3, and SOCS3. Scand. J. Med. Sci. Sports 2016, 26, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Koga, S.; Kojima, A.; Ishikawa, C.; Kuwabara, S.; Arai, K.; Yoshiyama, Y. Effects of Diet-Induced Obesity and Voluntary Exercise in a Tauopathy Mouse Model: Implications of Persistent Hyperleptinemia and Enhanced Astrocytic Leptin Receptor Expression. Neurobiol. Dis. 2014, 71, 180–192. [Google Scholar] [CrossRef] [PubMed]
- Perry, B.D.; Caldow, M.K.; Brennan-Speranza, T.C.; Sbaraglia, M.; Jerums, G.; Garnham, A.; Wong, C.; Levinger, P.; Haq, M.A.U.; Hare, D.L.; et al. Muscle Atrophy in Patients with Type 2 Diabetes Mellitus: Roles of Inflammatory Pathways, Physical Activity and Exercise. Exerc. Immunol. Rev. 2016, 22, 94–109. [Google Scholar] [PubMed]
- Parry, T.L.; Hayward, R. Exercise Protects against Cancer-Induced Cardiac Cachexia. Med. Sci. Sports Exerc. 2018, 50, 1169–1176. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, A.A.; Lira, F.S.; Pimentel, G.D.; de Souza, C.O.; Batatinha, H.; Biondo, L.A.; Yamashita, A.S.; Junior, E.A.; Neto, J.C. Aerobic Exercise Modulates the Free Fatty Acids and Inflammatory Response During Obesity and Cancer Cachexia. Crit. Rev. Eukaryot. Gene Expr. 2016, 26, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Maddocks, M.; Jones, L.W.; Wilcock, A. Immunological and Hormonal Effects of Exercise: Implications for Cancer Cachexia. Curr. Opin. Support Palliat. Care 2013, 7, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Lira, F.S.; Rosa, J.C.; Zanchi, N.E.; Yamashita, A.S.; Lopes, R.D.; Lopes, A.C.; Batista, M.L., Jr.; Seelaender, M. Regulation of Inflammation in the Adipose Tissue in Cancer Cachexia: Effect of Exercise. Cell Biochem. Funct. 2009, 27, 71–75. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, A.; Cheng, K.Y.; Gao, Y.; Seo, D.O.; Anton, S.; Carter, C.S.; Zhang, Y.; Tumer, N.; Scarpace, P.J. The Act of Voluntary Wheel Running Reverses Dietary Hyperphagia and Increases Leptin Signaling in Ventral Tegmental Area of Aged Obese Rats. Gerontology 2011, 57, 335–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patterson, C.M.; Bouret, S.G.; Dunn-Meynell, A.A.; Levin, B.E. Three Weeks of Postweaning Exercise in Dio Rats Produces Prolonged Increases in Central Leptin Sensitivity and Signaling. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2009, 296, R537–R548. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Jang, J.S.; Jun, D.W.; Hong, S.M. Exercise Enhances Insulin and Leptin Signaling in the Cerebral Cortex and Hypothalamus During Dexamethasone-Induced Stress in Diabetic Rats. Neuroendocrinology 2005, 82, 282–293. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Tian, Y.; Xu, J.; Liu, D.; Wang, X.; Zhao, B. Endurance Exercise Is a Leptin Signaling Mimetic in Hypothalamus of Wistar Rats. Lipids Health Dis. 2011, 10, 225. [Google Scholar] [CrossRef] [PubMed]
- Haddad, F.; Zaldivar, F.; Cooper, D.M.; Adams, G.R. IL-6-Induced Skeletal Muscle Atrophy. J. Appl. Physiol. (1985) 2005, 98, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.F.; Sanchez, B.J.; Hall, D.T.; Tremblay, A.K.; di Marco, S.; Gallouzi, I.E. STAT3 Promotes Ifngamma/Tnfalpha-Induced Muscle Wasting in an Nf-Kappab-Dependent and IL-6-Independent Manner. EMBO Mol. Med. 2017, 9, 622–637. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, S.D. Inhibition of STAT3 Signaling Ameliorates Atrophy of the Soleus Muscles in Mice Lacking the Vitamin D Receptor. Skelet. Muscle 2017, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Bonetto, A.; Aydogdu, T.; Jin, X.; Zhang, Z.; Zhan, R.; Puzis, L.; Koniaris, L.G.; Zimmers, T.A. Jak/STAT3 Pathway Inhibition Blocks Skeletal Muscle Wasting Downstream of IL-6 and in Experimental Cancer Cachexia. Am. J. Physiol. Endocrinol. Metab. 2012, 303, E410–E421. [Google Scholar] [CrossRef] [PubMed]
- Silva, K.A.; Dong, J.; Dong, Y.; Schor, N.; Tweardy, D.J.; Zhang, L.; Mitch, W.E. Inhibition of STAT3 Activation Suppresses Caspase-3 and the Ubiquitin-Proteasome System, Leading to Preservation of Muscle Mass in Cancer Cachexia. J. Biol. Chem. 2015, 290, 11177–11187. [Google Scholar] [CrossRef] [PubMed]
- Bonetto, A.; Aydogdu, T.; Kunzevitzky, N.; Guttridge, D.C.; Khuri, S.; Koniaris, L.G.; Zimmers, T.A. STAT3 Activation in Skeletal Muscle Links Muscle Wasting and the Acute Phase Response in Cancer Cachexia. PLoS ONE 2011, 6, e22538. [Google Scholar] [CrossRef] [PubMed]
- Ohara, R.; Fujita, Y.; Hata, K.; Nakagawa, M.; Yamashita, T. Axotomy Induces Axonogenesis in Hippocampal Neurons through STAT3. Cell Death Dis. 2011, 2, e175. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Neitzel, K.L.; Devlin, B.K.; MacLennan, A.J. STAT3 Phosphorylation in Injured Axons before Sensory and Motor Neuron Nuclei: Potential Role for STAT3 as a Retrograde Signaling Transcription Factor. J. Comp. Neurol. 2004, 474, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Natarajan, R.; Singal, V.; Benes, R.; Gao, J.; Chan, H.; Chen, H.; Yu, Y.; Zhou, J.; Wu, P. STAT3 Modulation to Enhance Motor Neuron Differentiation in Human Neural Stem Cells. PLoS ONE 2014, 9, e100405. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Shen, R.; Cho, H.H.; Kwon, R.J.; Seo, S.Y.; Lee, J.W.; Lee, S.K. STAT3 Promotes Motor Neuron Differentiation by Collaborating with Motor Neuron-Specific Lim Complex. Proc. Natl. Acad. Sci. USA 2013, 110, 11445–11450. [Google Scholar] [CrossRef] [PubMed]
- Miao, T.; Wu, D.; Zhang, Y.; Bo, X.; Subang, M.C.; Wang, P.; Richardson, P.M. Suppressor of Cytokine Signaling-3 Suppresses the Ability of Activated Signal Transducer and Activator of Transcription-3 to Stimulate Neurite Growth in Rat Primary Sensory Neurons. J. Neurosci. 2006, 26, 9512–9519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Snyder, M.; Huang, X.Y.; Zhang, J.J. Identification of Novel Direct STAT3 Target Genes for Control of Growth and Differentiation. J. Biol. Chem. 2008, 283, 3791–3798. [Google Scholar] [CrossRef] [PubMed]
- Vallania, F.; Schiavone, D.; Dewilde, S.; Pupo, E.; Garbay, S.; Calogero, R.; Pontoglio, M.; Provero, P.; Poli, V. Genome-Wide Discovery of Functional Transcription Factor Binding Sites by Comparative Genomics: The Case of STAT3. Proc. Natl. Acad. Sci. USA 2009, 106, 5117–5122. [Google Scholar] [CrossRef] [PubMed]
- Bourillot, P.Y.; Aksoy, I.; Schreiber, V.; Wianny, F.; Schulz, H.; Hummel, O.; Hubner, N.; Savatier, P. Novel STAT3 Target Genes Exert Distinct Roles in the Inhibition of Mesoderm and Endoderm Differentiation in Cooperation with Nanog. Stem Cells 2009, 27, 1760–1771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, Y.M.; Kim, J.K.; Choi, Y.; Choi, S.; Yoo, J.Y. Prediction and Experimental Validation of Novel STAT3 Target Genes in Human Cancer Cells. PLoS ONE 2009, 4, e6911. [Google Scholar] [CrossRef] [PubMed]
- Dauer, D.J.; Ferraro, B.; Song, L.; Yu, B.; Mora, L.; Buettner, R.; Enkemann, S.; Jove, R.; Haura, E.B. STAT3 Regulates Genes Common to Both Wound Healing and Cancer. Oncogene 2005, 24, 3397–3408. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xu, H.; Yuan, P.; Fang, F.; Huss, M.; Vega, V.B.; Wong, E.; Orlov, Y.L.; Zhang, W.; Jiang, J.; et al. Integration of External Signaling Pathways with the Core Transcriptional Network in Embryonic Stem Cells. Cell 2008, 133, 1106–1117. [Google Scholar] [CrossRef] [PubMed]
- Lieskovska, J.; Guo, D.; Derman, E. Growth Impairment in IL-6-Overexpressing Transgenic Mice Is Associated with Induction of SOCS3 mRNA. Growth Horm. IGF Res. 2003, 13, 26–35. [Google Scholar] [CrossRef]
- Dogra, C.; Srivastava, D.S.; Kumar, A. Protein-DNA Array-Based Identification of Transcription Factor Activities Differentially Regulated in Skeletal Muscle of Normal and Dystrophin-Deficient Mdx Mice. Mol. Cell. Biochem. 2008, 312, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xu, Y.; Li, W.; Wang, G.; Song, Y.; Yang, G.; Han, X.; Du, Z.; Sun, L.; Ma, K. STAT3 Induces Muscle Stem Cell Differentiation by Interaction with Myod. Cytokine 2009, 46, 137–141. [Google Scholar] [CrossRef] [PubMed]
- Burniston, J.G.; Kenyani, J.; Gray, D.; Guadagnin, E.; Jarman, I.H.; Cobley, J.N.; Cuthbertson, D.J.; Chen, Y.W.; Wastling, J.M.; Lisboa, P.J.; et al. Conditional Independence Mapping of DIGE Data Reveals PDIA3 Protein Species as Key Nodes Associated with Muscle Aerobic Capacity. J. Proteom. 2014, 106, 230–245. [Google Scholar] [CrossRef] [PubMed]
- Srikuea, R.; Esser, K.A.; Pholpramool, C. Leukaemia Inhibitory Factor Is Expressed in Rat Gastrocnemius Muscle after Contusion and Increases Proliferation of Rat L6 Myoblasts Via C-Myc Signalling. Clin. Exp. Pharmacol. Physiol. 2011, 38, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.H.; Choi, S.E.; Ha, E.S.; Jung, J.G.; Han, S.J.; Kim, H.J.; Kim, D.J.; Kang, Y.; Lee, K.W. IL-6 Induction of Tlr-4 Gene Expression via STAT3 Has an Effect on Insulin Resistance in Human Skeletal Muscle. Acta Diabetol. 2013, 50, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Megeney, L.A.; Perry, R.L.; LeCouter, J.E.; Rudnicki, M.A. bFGF and LIF Signaling Activates STAT3 in Proliferating Myoblasts. Dev. Genet. 1996, 19, 139–145. [Google Scholar] [CrossRef]
- Donohoe, C.L.; Ryan, A.M.; Reynolds, J.V. Cancer Cachexia: Mechanisms and Clinical Implications. Gastroenterol. Res. Pract. 2011, 2011, 601434. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, Y.; Hanna, D.L.; Zhang, W.; Baba, H.; Lenz, H.J. Molecular Pathways: Cachexia Signaling-a Targeted Approach to Cancer Treatment. Clin. Cancer Res. 2016, 22, 3999–4004. [Google Scholar] [CrossRef] [PubMed]
- Von Haehling, S.; Anker, S.D. Prevalence, Incidence and Clinical Impact of Cachexia: Facts and Numbers-Update 2014. J. Cachexia Sarcopenia Muscle 2014, 5, 261–263. [Google Scholar] [CrossRef] [PubMed]
- Tisdale, M.J. Mechanisms of Cancer Cachexia. Physiol. Rev. 2009, 89, 381–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baltgalvis, K.A.; Berger, F.G.; Pena, M.M.; Davis, J.M.; Muga, S.J.; Carson, J.A. Interleukin-6 and Cachexia in Apcmin/+ Mice. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2008, 294, R393–R401. [Google Scholar] [CrossRef] [PubMed]
- Strassmann, G.; Fong, M.; Kenney, J.S.; Jacob, C.O. Evidence for the Involvement of Interleukin 6 in Experimental Cancer Cachexia. J. Clin. Investig. 1992, 89, 1681–1684. [Google Scholar] [CrossRef] [PubMed]
- Soda, K.; Kawakami, M.; Kashii, A.; Miyata, M. Manifestations of Cancer Cachexia Induced by Colon 26 Adenocarcinoma Are Not Fully Ascribable to Interleukin-6. Int. J. Cancer 1995, 62, 332–336. [Google Scholar] [CrossRef] [PubMed]
- White, J.P.; Baltgalvis, K.A.; Puppa, M.J.; Sato, S.; Baynes, J.W.; Carson, J.A. Muscle Oxidative Capacity during IL-6-Dependent Cancer Cachexia. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 300, R201–R211. [Google Scholar] [CrossRef] [PubMed]
- Okada, S.; Okusaka, T.; Ishii, H.; Kyogoku, A.; Yoshimori, M.; Kajimura, N.; Yamaguchi, K.; Kakizoe, T. Elevated Serum Interleukin-6 Levels in Patients with Pancreatic Cancer. Jpn. J. Clin. Oncol. 1998, 28, 12–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebrahimi, B.; Tucker, S.L.; Li, D.; Abbruzzese, J.L.; Kurzrock, R. Cytokines in Pancreatic Carcinoma: Correlation with Phenotypic Characteristics and Prognosis. Cancer 2004, 101, 2727–2736. [Google Scholar] [CrossRef] [PubMed]
- Miura, T.; Mitsunaga, S.; Ikeda, M.; Shimizu, S.; Ohno, I.; Takahashi, H.; Furuse, J.; Inagaki, M.; Higashi, S.; Kato, H.; et al. Characterization of Patients with Advanced Pancreatic Cancer and High Serum Interleukin-6 Levels. Pancreas 2015, 44, 756–763. [Google Scholar] [CrossRef] [PubMed]
- Tamura, S.; Ouchi, K.F.; Mori, K.; Endo, M.; Matsumoto, T.; Eda, H.; Tanaka, Y.; Ishitsuka, H.; Tokita, H.; Yamaguchi, K. Involvement of Human Interleukin 6 in Experimental Cachexia Induced by a Human Uterine Cervical Carcinoma Xenograft. Clin. Cancer Res. 1995, 1, 1353–1358. [Google Scholar] [PubMed]
- Watchorn, T.M.; Waddell, I.; Dowidar, N.; Ross, J.A. Proteolysis-Inducing Factor Regulates Hepatic Gene Expression via the Transcription Factors NF-κB and STAT3. FASEB J. 2001, 15, 562–564. [Google Scholar] [CrossRef] [PubMed]
- Falconer, J.S.; Fearon, K.C.; Ross, J.A.; Elton, R.; Wigmore, S.J.; Garden, O.J.; Carter, D.C. Acute-Phase Protein Response and Survival Duration of Patients with Pancreatic Cancer. Cancer 1995, 75, 2077–2082. [Google Scholar] [CrossRef]
- Stephens, N.A.; Skipworth, R.J.; Fearon, K.C. Cachexia, Survival and the Acute Phase Response. Curr. Opin. Support. Palliat. Care 2008, 2, 267–274. [Google Scholar] [CrossRef] [PubMed]
- White, J.P.; Baynes, J.W.; Welle, S.L.; Kostek, M.C.; Matesic, L.E.; Sato, S.; Carson, J.A. The Regulation of Skeletal Muscle Protein Turnover During the Progression of Cancer Cachexia in the ApcMin/+ Mouse. PLoS ONE 2011, 6, e24650. [Google Scholar] [CrossRef] [PubMed]
- Mehl, K.A.; Davis, J.M.; Berger, F.G.; Carson, J.A. Myofiber Degeneration/Regeneration Is Induced in the Cachectic Apcmin/+ Mouse. J. Appl. Physiol. (1985) 2005, 99, 2379–2387. [Google Scholar] [CrossRef] [PubMed]
- He, W.A.; Berardi, E.; Cardillo, V.M.; Acharyya, S.; Aulino, P.; Thomas-Ahner, J.; Wang, J.; Bloomston, M.; Muscarella, P.; Nau, P.; et al. Nf-Kappab-Mediated Pax7 Dysregulation in the Muscle Microenvironment Promotes Cancer Cachexia. J. Clin. Investig. 2013, 123, 4821–4835. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, M.; de Rossi, M.; Barberi, L.; Musaro, A. IL-6 Impairs Myogenic Differentiation by Downmodulation of p90RSK/eEF2 and mTOR/p70S6K Axes, without Affecting AKT Activity. BioMed Res. Int. 2014, 2014, 206026. [Google Scholar] [CrossRef] [PubMed]
- Hillmer, E.J.; Zhang, H.; Li, H.S.; Watowich, S.S. STAT3 Signaling in Immunity. Cytokine Growth Factor Rev. 2016, 31, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Hansoul, S.; Nicolae, D.L.; Cho, J.H.; Duerr, R.H.; Rioux, J.D.; Brant, S.R.; Silverberg, M.S.; Taylor, K.D.; Barmada, M.M.; et al. Genome-Wide Association Defines More Than 30 Distinct Susceptibility Loci for Crohn’s Disease. Nat. Genet. 2008, 40, 955–962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, J.H. The Genetics and Immunopathogenesis of Inflammatory Bowel Disease. Nat. Rev. Immunol. 2008, 8, 458–466. [Google Scholar] [CrossRef] [PubMed]
- Jakkula, E.; Leppa, V.; Sulonen, A.M.; Varilo, T.; Kallio, S.; Kemppinen, A.; Purcell, S.; Koivisto, K.; Tienari, P.; Sumelahti, M.L.; et al. Genome-Wide Association Study in a High-Risk Isolate for Multiple Sclerosis Reveals Associated Variants in STAT3 Gene. Am. J. Hum. Genet. 2010, 86, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Tsoi, L.C.; Spain, S.L.; Knight, J.; Ellinghaus, E.; Stuart, P.E.; Capon, F.; Ding, J.; Li, Y.; Tejasvi, T.; Gudjonsson, J.E.; et al. Identification of 15 New Psoriasis Susceptibility Loci Highlights the Role of Innate Immunity. Nat. Genet. 2012, 44, 1341–1348. [Google Scholar] [CrossRef] [PubMed]
- Hilton-Jones, D. Observations on the Classification of the Inflammatory Myopathies. Presse Med. 2011, 40, e199–e208. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, F.; Guggino, G.; Rizzo, A.; Ferrante, A.; Raimondo, S.; Giardina, A.; Dieli, F.; Campisi, G.; Alessandro, R.; Triolo, G. Potential Involvement of IL-22 and IL-22-Producing Cells in the Inflamed Salivary Glands of Patients with Sjogren’s Syndrome. Ann. Rheum. Dis. 2012, 71, 295–301. [Google Scholar] [CrossRef] [PubMed]
- Ikeuchi, H.; Kuroiwa, T.; Hiramatsu, N.; Kaneko, Y.; Hiromura, K.; Ueki, K.; Nojima, Y. Expression of Interleukin-22 in Rheumatoid Arthritis: Potential Role as a Proinflammatory Cytokine. Arthritis Rheumatol. 2005, 52, 1037–1046. [Google Scholar] [CrossRef] [PubMed]
- Rutz, S.; Eidenschenk, C.; Ouyang, W. IL-22, Not Simply a Th17 Cytokine. Immunol. Rev. 2013, 252, 116–132. [Google Scholar] [CrossRef] [PubMed]
- Sterbank, J.; Marino, J.; Jhaveri, D.; Horbal, J.; Tcheurekdjian, H.; Hostoffer, R. A Unique Case of Peroneus Brevis/Longus Myositis in a Patient with a STAT3 Mutation. Ann. Allergy Asthma Immunol. 2013, 110, 204–205. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Canoves, P.; Scheele, C.; Pedersen, B.K.; Serrano, A.L. Interleukin-6 Myokine Signaling in Skeletal Muscle: A Double-Edged Sword? FEBS J. 2013, 280, 4131–4148. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, L.; Berardinelli, M.G.; de Pasquale, L.; Nicoletti, C.; D’Amico, A.; Carvello, F.; Moneta, G.M.; Catizone, A.; Bertini, E.; de Benedetti, F.; et al. Functional and Morphological Improvement of Dystrophic Muscle by Interleukin 6 Receptor Blockade. EBioMedicine 2015, 2, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Danieli-Betto, D.; Peron, S.; Germinario, E.; Zanin, M.; Sorci, G.; Franzoso, S.; Sandona, D.; Betto, R. Sphingosine 1-Phosphate Signaling Is Involved in Skeletal Muscle Regeneration. Am. J. Physiol. Cell Physiol. 2010, 298, C550–C558. [Google Scholar] [CrossRef] [PubMed]
- Nagata, Y.; Partridge, T.A.; Matsuda, R.; Zammit, P.S. Entry of Muscle Satellite Cells into the Cell Cycle Requires Sphingolipid Signaling. J. Cell Biol. 2006, 174, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Loh, K.C.; Leong, W.I.; Carlson, M.E.; Oskouian, B.; Kumar, A.; Fyrst, H.; Zhang, M.; Proia, R.L.; Hoffman, E.P.; Saba, J.D. Sphingosine-1-Phosphate Enhances Satellite Cell Activation in Dystrophic Muscles through a S1PR2/STAT3 Signaling Pathway. PLoS ONE 2012, 7, e37218. [Google Scholar] [CrossRef]
- Willmann, R.; Possekel, S.; Dubach-Powell, J.; Meier, T.; Ruegg, M.A. Mammalian Animal Models for Duchenne Muscular Dystrophy. Neuromuscul. Disord. 2009, 19, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Deconinck, A.E.; Rafael, J.A.; Skinner, J.A.; Brown, S.C.; Potter, A.C.; Metzinger, L.; Watt, D.J.; Dickson, J.G.; Tinsley, J.M.; Davies, K.E. Utrophin-Dystrophin-Deficient Mice as a Model for Duchenne Muscular Dystrophy. Cell 1997, 90, 717–727. [Google Scholar] [CrossRef] [Green Version]
- Nakamori, M.; Hamanaka, K.; Thomas, J.D.; Wang, E.T.; Hayashi, Y.K.; Takahashi, M.P.; Swanson, M.S.; Nishino, I.; Mochizuki, H. Aberrant Myokine Signaling in Congenital Myotonic Dystrophy. Cell Rep. 2017, 21, 1240–1252. [Google Scholar] [CrossRef] [PubMed]
- Janssen, S.P.; Gayan-Ramirez, G.; van den Bergh, A.; Herijgers, P.; Maes, K.; Verbeken, E.; Decramer, M. Interleukin-6 Causes Myocardial Failure and Skeletal Muscle Atrophy in Rats. Circulation 2005, 111, 996–1005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massague, J. Tgfbeta Signalling in Context. Nat. Rev. Mol. Cell Biol. 2012, 13, 616–630. [Google Scholar] [CrossRef] [PubMed]
- Leask, A.; Abraham, D.J. TGF-β Signaling and the Fibrotic Response. FASEB J. 2004, 18, 816–827. [Google Scholar] [CrossRef] [PubMed]
- Annes, J.P.; Munger, J.S.; Rifkin, D.B. Making Sense of Latent TGFβ Activation. J. Cell Sci. 2003, 116, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Pohlers, D.; Brenmoehl, J.; Loffler, I.; Muller, C.K.; Leipner, C.; Schultze-Mosgau, S.; Stallmach, A.; Kinne, R.W.; Wolf, G. TGF-β and Fibrosis in Different Organs—Molecular Pathway Imprints. Biochim. Biophys. Acta 2009, 1792, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Attisano, L.; Wrana, J.L. Signal Transduction by the TGF-β Superfamily. Science 2002, 296, 1646–1647. [Google Scholar] [CrossRef] [PubMed]
- Cencetti, F.; Bernacchioni, C.; Tonelli, F.; Roberts, E.; Donati, C.; Bruni, P. TGFβ1 Evokes Myoblast Apoptotic Response via a Novel Signaling Pathway Involving S1P4 Transactivation Upstream of Rho-Kinase-2 Activation. FASEB J. 2013, 27, 4532–4546. [Google Scholar] [CrossRef] [PubMed]
- Narola, J.; Pandey, S.N.; Glick, A.; Chen, Y.W. Conditional Expression of TGF-β1 in Skeletal Muscles Causes Endomysial Fibrosis and Myofibers Atrophy. PLoS ONE 2013, 8, e79356. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Foster, W.; Deasy, B.M.; Chan, Y.; Prisk, V.; Tang, Y.; Cummins, J.; Huard, J. Transforming Growth Factor-Beta1 Induces the Differentiation of Myogenic Cells into Fibrotic Cells in Injured Skeletal Muscle: A Key Event in Muscle Fibrogenesis. Am. J. Pathol. 2004, 164, 1007–1019. [Google Scholar] [CrossRef]
- Ceco, E.; McNally, E.M. Modifying Muscular Dystrophy through Transforming Growth Factor-Beta. FEBS J. 2013, 280, 4198–4209. [Google Scholar] [CrossRef] [PubMed]
- Lieber, R.L.; Ward, S.R. Cellular Mechanisms of Tissue Fibrosis. 4. Structural and Functional Consequences of Skeletal Muscle Fibrosis. Am. J. Physiol. Cell Physiol. 2013, 305, C241–C252. [Google Scholar] [CrossRef] [PubMed]
- Lefaucheur, J.P.; Sebille, A. Muscle Regeneration Following Injury Can Be Modified in Vivo by Immune Neutralization of Basic Fibroblast Growth Factor, Transforming Growth Factor Beta 1 or Insulin-Like Growth Factor I. J. Neuroimmunol. 1995, 57, 85–91. [Google Scholar] [CrossRef]
- Ogata, H.; Chinen, T.; Yoshida, T.; Kinjyo, I.; Takaesu, G.; Shiraishi, H.; Iida, M.; Kobayashi, T.; Yoshimura, A. Loss of SOCS3 in the Liver Promotes Fibrosis by Enhancing STAT3-Mediated TGF-β1 Production. Oncogene 2006, 25, 2520–2530. [Google Scholar] [CrossRef] [PubMed]
- Bromberg, J.; Darnell, J.E., Jr. The Role of STATs in Transcriptional Control and Their Impact on Cellular Function. Oncogene 2000, 19, 2468–2473. [Google Scholar] [CrossRef] [PubMed]
- Bromberg, J.F.; Wrzeszczynska, M.H.; Devgan, G.; Zhao, Y.; Pestell, R.G.; Albanese, C.; Darnell, J.E., Jr. STAT3 as an Oncogene. Cell 1999, 98, 295–303. [Google Scholar] [CrossRef]
- Wang, G.; Yu, Y.; Sun, C.; Liu, T.; Liang, T.; Zhan, L.; Lin, X.; Feng, X.H. STAT3 Selectively Interacts with Smad3 to Antagonize TGF-β Signalling. Oncogene 2016, 35, 4422. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Le, X.; Zheng, L.; Wang, L.; Frey, J.A.; Gao, A.C.; Peng, Z.; Huang, S.; Xiong, H.Q.; Abbruzzese, J.L.; et al. STAT3 Activation Regulates the Expression of Vascular Endothelial Growth Factor and Human Pancreatic Cancer Angiogenesis and Metastasis. Oncogene 2003, 22, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Presser, L.D.; McRae, S.; Waris, G. Activation of TGF-β1 Promoter by Hepatitis C Virus-Induced AP-1 and Sp1: Role of TGF-β1 in Hepatic Stellate Cell Activation and Invasion. PLoS ONE 2013, 8, e56367. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.Y.; Heller, M.; Meng, Z.; Yu, L.R.; Tang, Y.; Zhou, M.; Zhang, Y.E. Transforming Growth Factor-Beta (TGF-β) Directly Activates the Jak1-STAT3 Axis to Induce Hepatic Fibrosis in Coordination with the Smad Pathway. J. Biol. Chem. 2017, 292, 4302–4312. [Google Scholar] [CrossRef] [PubMed]
- Li, S.W.; Wang, C.Y.; Jou, Y.J.; Yang, T.C.; Huang, S.H.; Wan, L.; Lin, Y.J.; Lin, C.W. SARS Coronavirus Papain-Like Protease Induces Egr-1-Dependent up-Regulation of TGF-β1 via ROS/P38 MAPK/STAT3 Pathway. Sci. Rep. 2016, 6, 25754. [Google Scholar] [CrossRef] [PubMed]

| Gene Symbol | Regulatory Target | Reference |
|---|---|---|
| c-Fos | Direct | Trenerry et al., 2007 [23] |
| Socs3 | Direct | Lieskovska et al., 2003 [56] |
| Jun | Direct | Dogra et al., 2008 [57] |
| Myod1 | Direct | Yang et al., 2009 [58] |
| Pdia3 | Direct | Burniston et al., 2014 [59] |
| c-Myc | Direct | Srikuea et al., 2011 [60] |
| Tlr4 | Indirect | Kim et al., 2013 [61] |
| Lif | ND | Megeney et al., 1996 [62] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guadagnin, E.; Mázala, D.; Chen, Y.-W. STAT3 in Skeletal Muscle Function and Disorders. Int. J. Mol. Sci. 2018, 19, 2265. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19082265
Guadagnin E, Mázala D, Chen Y-W. STAT3 in Skeletal Muscle Function and Disorders. International Journal of Molecular Sciences. 2018; 19(8):2265. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19082265
Chicago/Turabian StyleGuadagnin, Eleonora, Davi Mázala, and Yi-Wen Chen. 2018. "STAT3 in Skeletal Muscle Function and Disorders" International Journal of Molecular Sciences 19, no. 8: 2265. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19082265