Genome-Wide Bioinformatics Analysis of MAPK Gene Family in Kiwifruit (Actinidia Chinensis)
Abstract
:1. Introduction
2. Results
2.1. Identification of MAPK Family Genes in Kiwifruit
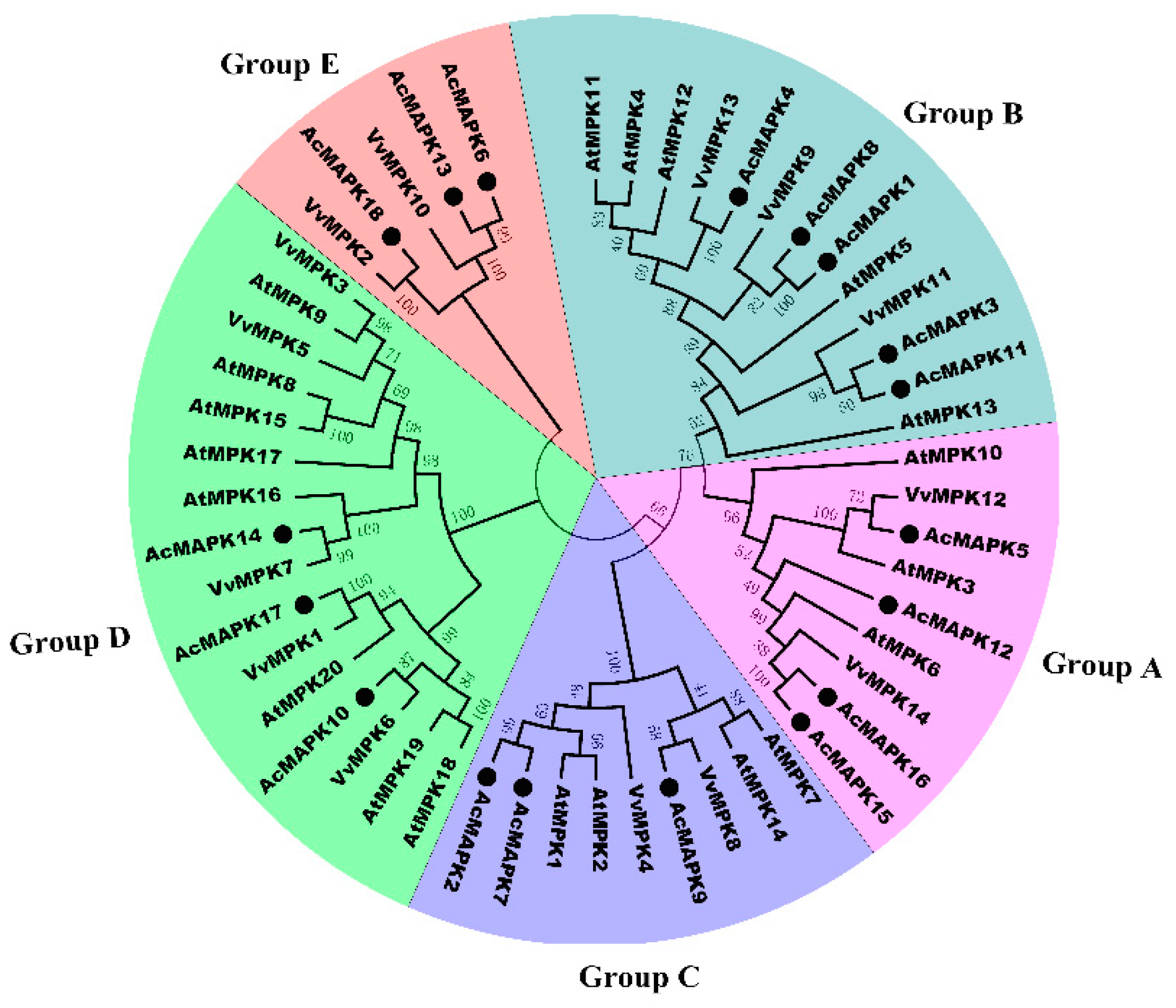
2.2. Phylogenetic Relationship Analysis of MAPK Gene in Kiwifruit
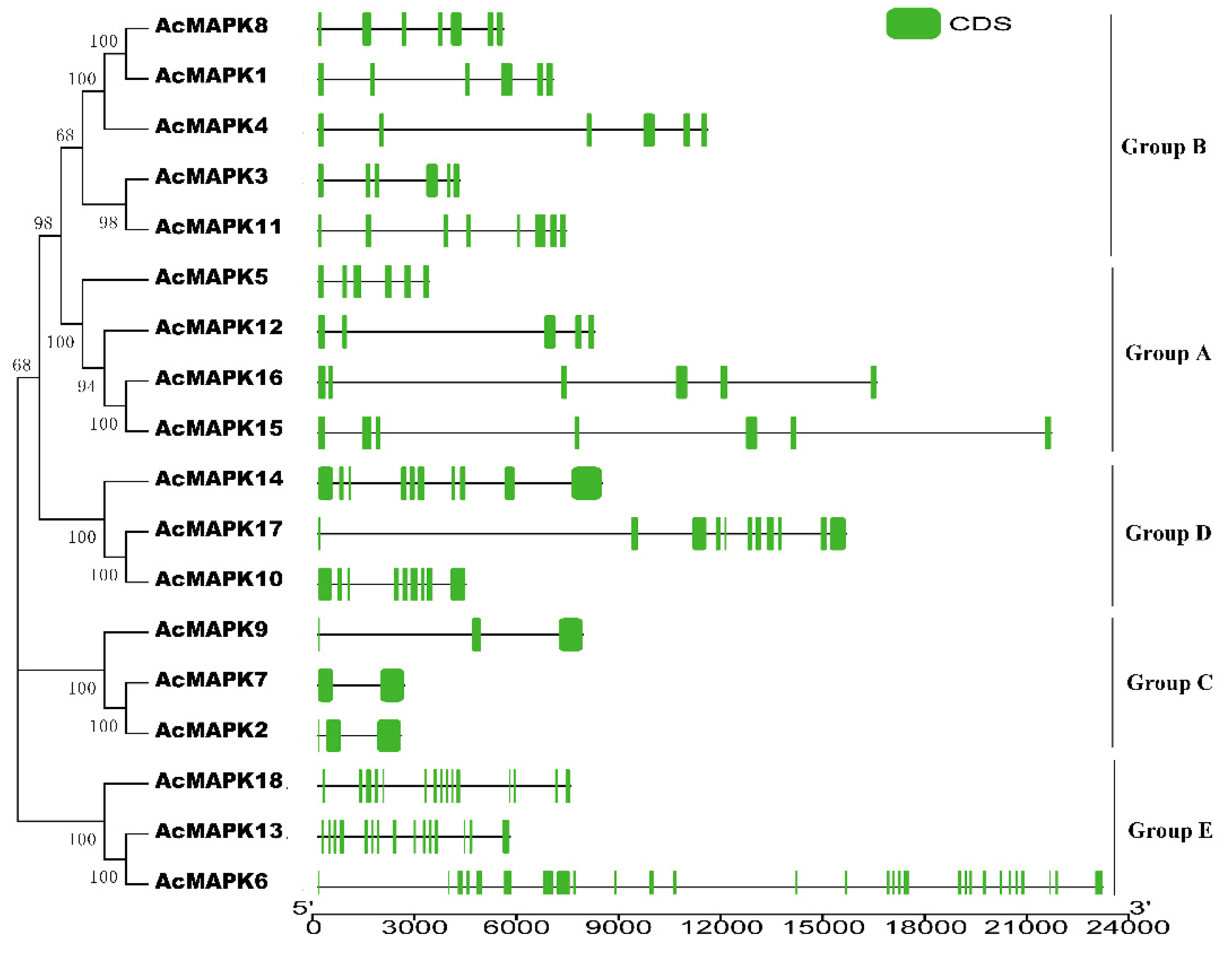
2.3. Gene Structure Analysis of MAPK Gene in Kiwifruit
2.4. The Conserved Motifs Domain and Promoter Regions Analysis of MAPK Gene in Kiwifruit
2.5. Expression Profiles of AcMAPK Genes in Response to Hormone Treatments
2.6. Expression Patterns of AcMAPK Genes under Abiotic and Biotic Stresses
3. Discussion
4. Materials and Methods
4.1. Genome-Wide Identification of MAPK Genes in Kiwifruit
4.2. Sequence Alignment, Phylogenetic Analysis, Chromosomal Location, and Gene Structure Construction
4.3. Cis-Element Analysis of Putative Promoter Regions
4.4. Plant Materials and Treatments
4.5. Total RNA Isolation and qRT-PCR Expression Analysis
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Hamel, L.P.; Nicole, M.C.; Sritubtim, S.; Morency, M.J.; Ellis, M.; Ehlting, J.; Beaudoin, N.; Barbazuk, B.; Klessig, D. Ancient signals: Comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 2006, 11, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Romeis, T. Protein kinases in the plant defence response. Curr. Opin. Plant Biol. 2001, 4, 407–414. [Google Scholar] [CrossRef]
- Group, M. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 2002, 7, 301–308. [Google Scholar]
- Rodriguez, M.C.; Petersen, M.; Mundy, J. Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 2010, 61, 621–649. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Lovato, A.; Polverari, A.; Wang, M.; Liang, Y.H.; Ma, Y.C.; Cheng, Z.M. Genome-wide identification and analysis of mitogen activated protein kinase kinase kinase gene family in grapevine (Vitis vinifera). BMC Plant Biol. 2014, 14, 219. [Google Scholar] [CrossRef] [PubMed]
- Jonak, C.; Okresz, L.; Bogre, L.; Hirt, H. Complexity, cross talk and integration of plant MAP kinase signalling. Curr. Opin. Plant Biol. 2002, 5, 415–424. [Google Scholar] [CrossRef]
- Pitzschke, A.; Schikora, A.; Hirt, H. MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 2009, 12, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Champion, A.; Picaud, A.; Henry, Y. Reassessing the MAP3K and MAP4K relationships. Trends Plant Sci. 2004, 9, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Mohanta, T.K.; Arora, P.K.; Mohanta, N.; Parida, P.; Bae, H. Identification of new members of the MAPK gene family in plants shows diverse conserved domains and novel activation loop variants. BMC Genom. 2015, 16, 58. [Google Scholar] [CrossRef] [PubMed]
- Doczi, R.; Okresz, L.; Romero, A.E.; Paccanaro, A.; Bogre, L. Exploring the evolutionary path of plant MAPK networks. Trends Plant Sci. 2012, 17, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Colcombet, J.; Hirt, H. Arabidopsis MAPKs: A complex signalling network involved in multiple biological processes. Biochem. J. 2008, 413, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.S.; Luo, T.; Fu, X.Z.; Fan, Q.J.; Liu, J.H. Cloning and molecular characterization of a mitogen-activated protein kinase gene from Poncirus trifoliata whose ectopic expression confers dehydration/drought tolerance in transgenic tobacco. J. Exp. Bot. 2011, 62, 5191–5206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Lovato, A.; Liang, Y.H.; Wang, M.; Chen, F.; Tornielli, G.B.; Polverari, A.; Pezzotti, M.; Cheng, Z.M. Validation by isolation and expression analyses of MAPK gene family in grapevine (Vitis vinifera L.). Aust. J. Grape Wine Res. 2014, 2, 255–262. [Google Scholar] [CrossRef]
- Reyna, N.S.; Yang, Y. Molecular analysis of the rice MAP kinase gene family in relation to Magnaporthe grisea infection. Mol. Plant Microbe Interact. 2006, 19, 530–540. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Wang, J.; Cheng, L.; Liu, S.; Wu, J.; Peng, Z.; Lu, G. Genome-wide analysis of the mitogen-activated protein kinase gene family in Solanum lycopersicum. Gene 2012, 499, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Pan, J.; Zhang, D.; Jiang, S.; Cai, G.; Wang, L.; Li, D. Identification of mitogen-activated protein kinase kinase gene family and MKK-MAPK interaction network in maize. Biochem. Biophys. Res. Commun. 2013, 441, 964–969. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hu, W.; Tan, S.; Wang, M.; Ma, Z.; Zhou, S.; Deng, X.; Zhang, Y.; Huang, C.; Yang, G. Genome-wide identification and analysis of MAPK and MAPKK gene families in Brachypodium distachyon. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Cakir, B.; Kilickaya, O. Mitogen-activated protein kinase cascades in Vitis vinifera. Front. Plant Sci. 2015, 6, 556. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Ren, S.; Han, Y.; Zhang, Q.; Qin, L.; Xing, Y. Identification and Analysis of Mitogen-Activated Protein Kinase (MAPK) Cascades in Fragaria vesca. Int. J. Mol. Sci. 2017, 18, 1766. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Cheng, T.; Wang, G.; Yan, Y.; Xia, Q. Cloning and evolutionary analysis of tobacco MAPK gene family. Mol. Biol. Rep. 2013, 40, 1407–1415. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Liu, X.; Long, D.; Guo, Q.; Fang, Y.; Bian, C.; Zhang, D.; Zeng, Q.; Xiang, Z.; Zhao, A. Molecular cloning and expression analysis of mulberry MAPK gene family. Plant Physiol. Biochem. 2014, 77, 108–116. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Guo, W.; Lu, J.; Yu, H.; Qu, C.; Tang, Z.; Li, J.; Chai, Y.; Liang, Y. Genome-Wide Survey and Expression Profile Analysis of the Mitogen-Activated Protein Kinase (MAPK) Gene Family in Brassica rapa. PLoS ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Nicole, M.C.; Hamel, L.P.; Morency, M.J.; Beaudoin, N.; Ellis, B.E.; Seguin, A. MAP-ping genomic organization and organ-specific expression profiles of poplar MAP kinases and MAP kinase kinases. BMC Genom. 2006, 7, 223. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Xu, R.; Luo, X.; Jiang, Z.; Shu, H. Genome-wide identification and expression analysis of MAPK and MAPKK gene family in Malus domestica. Gene 2013, 531, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Asai, T.; Tena, G.; Plotnikova, J.; Willmann, M.R.; Chiu, W.L.; Gomez-Gomez, L.; Boller, T.; Ausubel, F.M.; Sheen, J. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 2002, 415, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Galletti, R.; Ferrari, S.; De Lorenzo, G. Arabidopsis MPK3 and MPK6 play different roles in basal and oligogalacturonide- or flagellin-induced resistance against Botrytis cinerea. Plant Physiol. 2011, 157, 804–814. [Google Scholar] [CrossRef] [PubMed]
- Bush, S.M.; Krysan, P.J. Mutational evidence that the Arabidopsis MAP kinase MPK6 is involved in anther, inflorescence, and embryo development. J. Exp. Bot. 2007, 58, 2181–2191. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.E.; Hetherington, A.M. Plant development: YODA the stomatal switch. Curr. Biol. 2004, 14, 488–490. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.; Brodersen, P.; Naested, H.; Andreasson, E.; Lindhart, U.; Johansen, B.; Nielsen, H.B.; Lacy, M.; Austin, M.J.; Parker, J.E. Arabidopsis map kinase 4 negatively regulates systemic acquired resistance. Cell 2000, 103, 1111–1120. [Google Scholar] [CrossRef]
- Qiu, J.L.; Zhou, L.; Yun, B.W.; Nielsen, H.B.; Fiil, B.K.; Petersen, K.; Mackinlay, J.; Loake, G.J.; Mundy, J.; Morris, P.C. Arabidopsis mitogen-activated protein kinase kinases MKK1 and MKK2 have overlapping functions in defense signaling mediated by MEKK1, MPK4, and MKS1. Plant Physiol. 2008, 148, 212–222. [Google Scholar] [CrossRef] [PubMed]
- Teige, M.; Scheikl, E.; Eulgem, T.; Doczi, R.; Ichimura, K.; Shinozaki, K.; Dangl, J.L.; Hirt, H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 2004, 15, 141–152. [Google Scholar] [CrossRef] [PubMed]
- Kishi-Kaboshi, M.; Okada, K.; Kurimoto, L.; Murakami, S.; Umezawa, T.; Shibuya, N.; Yamane, H.; Miyao, A.; Takatsuji, H.; Takahashi, A.; et al. A rice fungal MAMP-responsive MAPK cascade regulates metabolic flow to antimicrobial metabolite synthesis. Plant J. 2010, 63, 599–612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.K.; Kim, B.G.; Kwon, T.R.; Jeong, M.J.; Park, S.R.; Lee, J.W.; Byun, M.O.; Kwon, H.B.; Matthews, B.F.; Hong, C.B.; et al. Overexpression of the mitogen-activated protein kinase gene OsMAPK33 enhances sensitivity to salt stress in rice (Oryza sativa L.). J. Biosci. 2011, 36, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Ding, H.; Zhang, A.; Ma, F.; Cao, J.; Jiang, M. A novel mitogen-activated protein kinase gene in maize (Zea mays), ZmMPK3, is involved in response to diverse environmental cues. J. Integr. Plant Biol. 2010, 52, 442–452. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Zhang, J.; Ye, N.; Cao, J.; Tan, M.; Zhang, J.; Jiang, M. ZmMPK5 is required for the NADPH oxidase-mediated self-propagation of apoplastic H2O2 in brassinosteroid-induced antioxidant defence in leaves of maize. J. Exp. Bot. 2010, 61, 4399–4411. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Zhang, M.; Kong, X.; Xing, X.; Liu, Y.; Zhou, Y.; Liu, Y.; Sun, L.; Li, D. ZmMPK17, a novel maize group D MAP kinase gene, is involved in multiple stress responses. Planta 2012, 235, 661–676. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Mao, H.; Dong, C.; Ji, R.; Cai, L.; Fu, H.; Liu, S. Overexpression of Brassica napus MPK4 enhances resistance to Sclerotinia sclerotiorum in oilseed rape. Mol. Plant Microbe Interact. 2009, 22, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; An, H.L.; Zhang, L.; Gao, Z.; Guo, X.Q. GhMPK7, a novel multiple stress-responsive cotton group C MAPK gene, has a role in broad spectrum disease resistance and plant development. Plant Mol. Biol. 2010, 74, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Ding, Z.; Ruan, M.; Yu, X.; Peng, M.; Liu, Y. Kiwifruit R2R3-MYB transcription factors and contribution of the novel AcMYB75 to red kiwifruit anthocyanin biosynthesis. Sci. Rep. 2017, 7, 16861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, S.; Ding, J.; Deng, D.; Tang, W.; Sun, H.; Liu, D.; Zhang, L.; Niu, X.; Zhang, X.; Meng, M.; et al. Draft genome of the kiwifruit Actinidia chinensis. Nat. Commun. 2013, 4, 2640. [Google Scholar] [CrossRef] [PubMed]
- Nonis, A.; Ruperti, B.; Pierasco, A.; Canaguier, A.; Adam-Blondon, A.F.; Di Gaspero, G.; Vizzotto, G. Neutral invertases in grapevine and comparative analysis with Arabidopsis, poplar and rice. Planta 2008, 229, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.R.; Kirti, P.B. A mitogen-activated protein kinase, AhMPK6 from peanut localizes to the nucleus and also induces defense responses upon transient expression in tobacco. Plant Physiol. Biochem. 2010, 48, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Tohge, T.; Fernie, A.R. Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nat. Protoc. 2010, 5, 1210–1227. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Masia, D.; Perez-Amador, M.A.; Carbonell, J.; Marcote, M.J. Diverse stress signals activate the C1 subgroup MAP kinases of Arabidopsis. FEBS Lett. 2007, 581, 1834–1840. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.; O’Hely, M.; Walsh, B.; Force, A. The probability of preservation of a newly arisen gene duplicate. Genetics 2001, 159, 1789–1804. [Google Scholar] [PubMed]
- Takahashi, F.; Yoshida, R.; Ichimura, K.; Mizoguchi, T.; Seo, S.; Yonezawa, M.; Maruyama, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The mitogen-activated protein kinase cascade MKK3-MPK6 is an important part of the jasmonate signal transduction pathway in Arabidopsis. Plant Cell 2007, 19, 805–818. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Zhang, S. MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 2013, 51, 245–266. [Google Scholar] [CrossRef] [PubMed]
- Meldau, S.; Ullman-Zeunert, L.; Govind, G.; Bartram, S.; Baldwin, I.T. MAPK-dependent JA and SA signalling in Nicotiana attenuata affects plant growth and fitness during competition with conspecifics. BMC Plant Biol. 2012, 12, 213. [Google Scholar] [CrossRef] [PubMed]
- Doczi, R.; Brader, G.; Pettko-Szandtner, A.; Rajh, I.; Djamei, A.; Pitzschke, A.; Teige, M.; Hirt, H. The Arabidopsis mitogen-activated protein kinase kinase MKK3 is upstream of group C mitogen-activated protein kinases and participates in pathogen signaling. Plant Cell 2007, 19, 3266–3279. [Google Scholar] [CrossRef] [PubMed]
- Hung, J.H.; Weng, Z. Sequence Alignment and Homology Search with BLAST and ClustalW. Cold Spring Harb. Protoc. 2016, 2016. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.Y.; Zhu, Q.H.; Chen, X.; Luo, J.C. GSDS: A gene structure display server. Yi Chuan 2007, 29, 1023–1026. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Pan, C.; Wang, Y.; Ye, L.; Wu, J.; Chen, L.; Zou, T.; Lu, G. Genome-wide identification of MAPK, MAPKK, and MAPKKK gene families and transcriptional profiling analysis during development and stress response in cucumber. BMC Genom. 2015, 16, 386. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Huang, S.N.; Wang, G.; Xuan, J.P.; Guo, Z.R. Overexpression of Actinidia deliciosa pyruvate decarboxylase 1 gene enhances waterlogging stress in transgenic Arabidopsis thaliana. Plant Physiol. Biochem. 2016, 106, 244–252. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.R.; Allan, A.C.; Chen, K.S.; Ferguson, I.B. Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiol. 2010, 153, 1280–1292. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−∆∆Ct) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



| Name | Gene ID | Chromosome | Length of Protein in AA (Amino Acid) | CDS (Coding Sequences) Length in bp | MW (Molecular Weights) (kDa) | PI (Protein Isoelectric Points) | Number of Exons | T-Loop | Subcellular Location |
|---|---|---|---|---|---|---|---|---|---|
| AcMAPK1 | Achn005721 | 15 | 374 | 1125 | 42.93 | 6.44 | 6 | TEY | Chloroplast |
| AcMAPK2 | Achn025711 | 11 | 379 | 1140 | 43.65 | 7.12 | 3 | TEY | Nuclear, Cytoplasm |
| AcMAPK3 | Achn060571 | 28 | 340 | 1023 | 38.99 | 4.89 | 6 | TEY | Cytoplasm |
| AcMAPK4 | Achn074341 | Un | 376 | 1131 | 43.36 | 6.51 | 6 | TEY | Cytoplasm |
| AcMAPK5 | Achn082251 | 23 | 344 | 1035 | 39.74 | 5.41 | 6 | TEY | Cytoplasm |
| AcMAPK6 | Achn098501 | 20 | 1056 | 3171 | 119.46 | 9.14 | 29 | TEY | Cytoplasm |
| AcMAPK7 | Achn131961 | Un | 371 | 1116 | 42.56 | 6.89 | 2 | TEY | Cytoplasm |
| AcMAPK8 | Achn132381 | 25 | 434 | 1305 | 49.42 | 7.39 | 7 | TEY | Cytoplasm |
| AcMAPK9 | Achn135551 | 1 | 336 | 1011 | 39.11 | 7.90 | 3 | TEY | Peroxisome |
| AcMAPK10 | Achn146591 | 13 | 601 | 1806 | 67.76 | 9.17 | 9 | TDY | Chloroplast |
| AcMAPK11 | Achn195331 | 13 | 425 | 1278 | 48.30 | 4.52 | 8 | TEY | Vacuolar |
| AcMAPK12 | Achn209161 | 1 | 340 | 1023 | 38.82 | 6.25 | 5 | TEY | Cytoplasm |
| AcMAPK13 | Achn228801 | 25 | 451 | 1356 | 51.08 | 9.82 | 16 | TEY | Cytoplasm, Nuclear |
| AcMAPK14 | Achn237151 | 29 | 862 | 2589 | 97.02 | 9.25 | 10 | TDY | Nuclear, Chloroplast |
| AcMAPK15 | Achn248791 | Un | 475 | 1428 | 54.89 | 5.15 | 7 | TEY | Cytoplasm |
| AcMAPK16 | Achn252431 | 2 | 403 | 1212 | 46.22 | 5.69 | 6 | TEY | Cytoplasm |
| AcMAPK17 | Achn296271 | 1 | 702 | 2109 | 79.02 | 9.03 | 11 | TDY | Chloroplast |
| AcMAPK18 | Achn377281 | 15 | 409 | 1230 | 46.34 | 5.40 | 16 | TDY | Nuclear |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, G.; Wang, T.; Jia, Z.-H.; Xuan, J.-P.; Pan, D.-L.; Guo, Z.-R.; Zhang, J.-Y. Genome-Wide Bioinformatics Analysis of MAPK Gene Family in Kiwifruit (Actinidia Chinensis). Int. J. Mol. Sci. 2018, 19, 2510. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19092510
Wang G, Wang T, Jia Z-H, Xuan J-P, Pan D-L, Guo Z-R, Zhang J-Y. Genome-Wide Bioinformatics Analysis of MAPK Gene Family in Kiwifruit (Actinidia Chinensis). International Journal of Molecular Sciences. 2018; 19(9):2510. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19092510
Chicago/Turabian StyleWang, Gang, Tao Wang, Zhan-Hui Jia, Ji-Ping Xuan, De-Lin Pan, Zhong-Ren Guo, and Ji-Yu Zhang. 2018. "Genome-Wide Bioinformatics Analysis of MAPK Gene Family in Kiwifruit (Actinidia Chinensis)" International Journal of Molecular Sciences 19, no. 9: 2510. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19092510






