Identification of Novel Quantitative Trait Loci Linked to Crown Rot Resistance in Spring Wheat
Abstract
:1. Introduction
2.Results
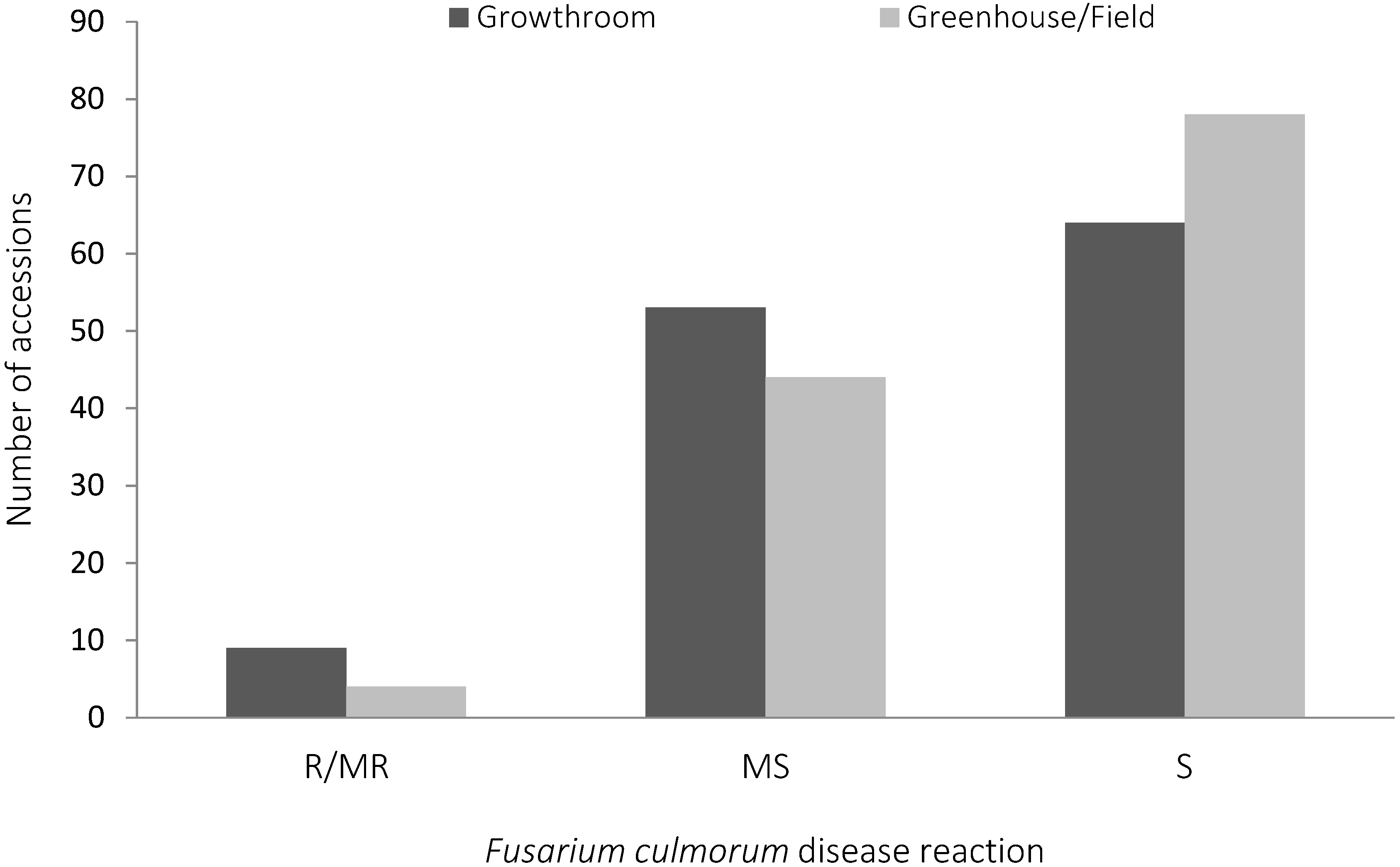
2.1. Fusarium culmorum Disease Assessment
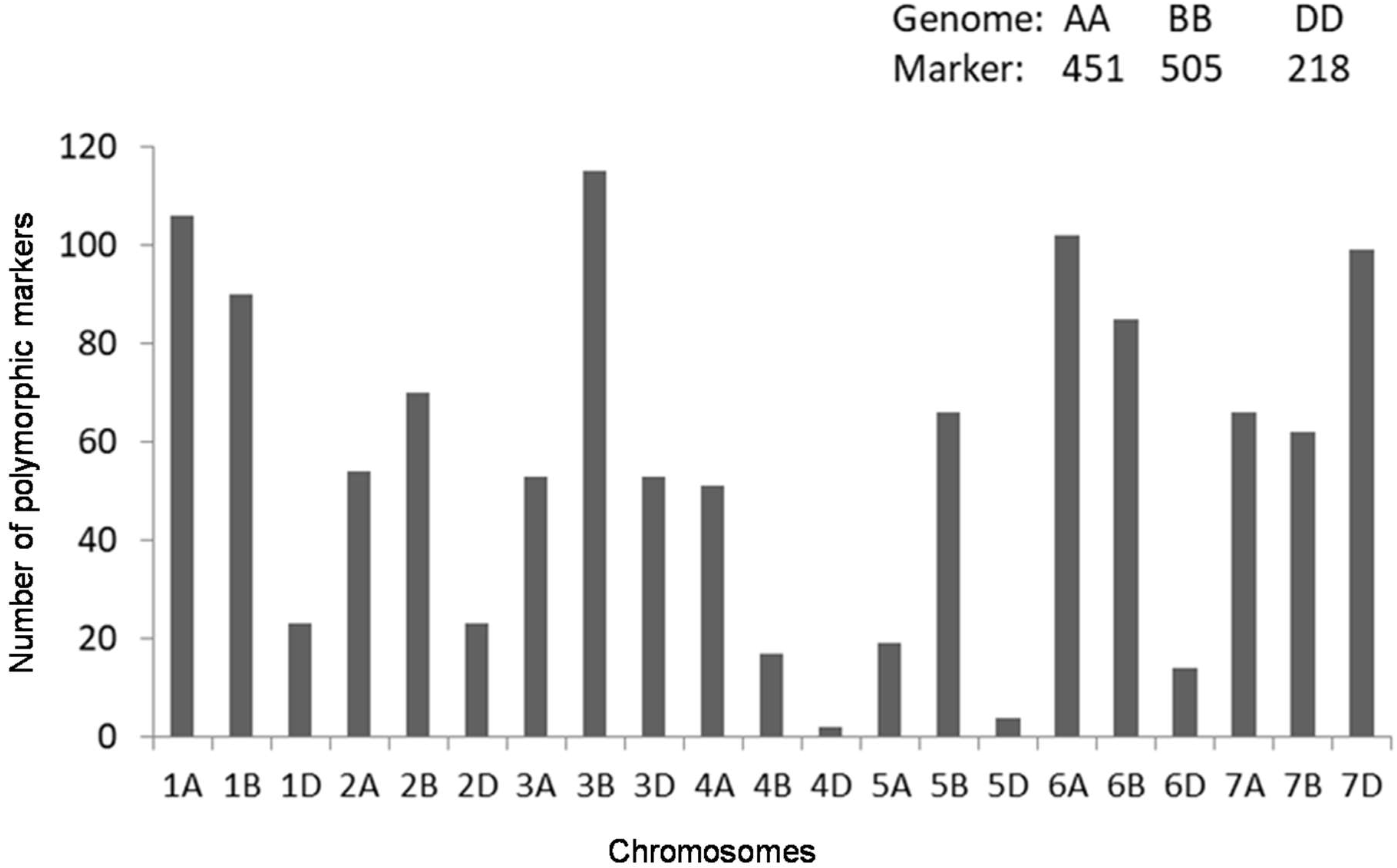
2.2. Genotyping with Diversity Arrays Technology (DArT)Markers
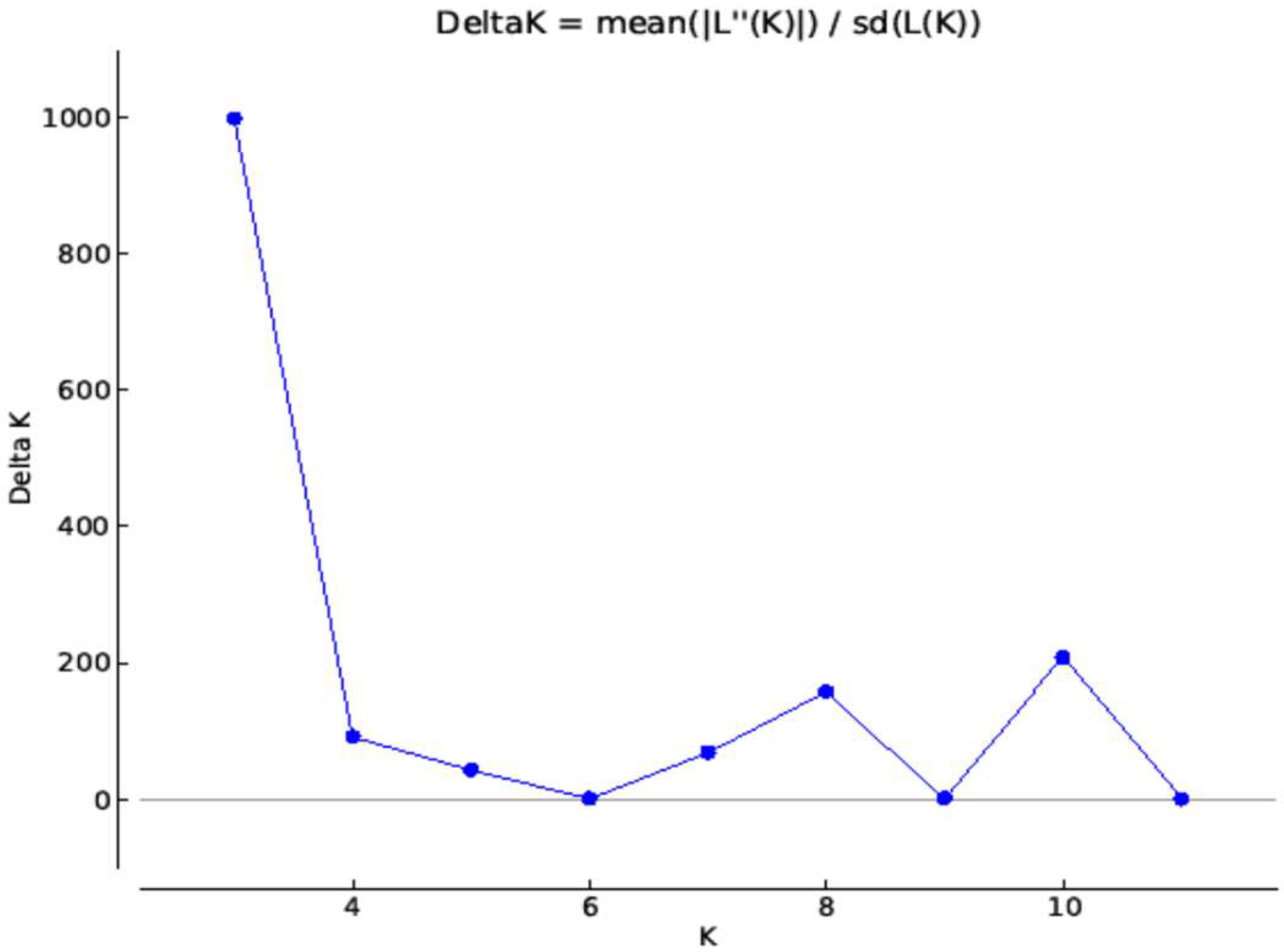
2.3. Structure Analysis
2.4. Linkage Disequilibrium
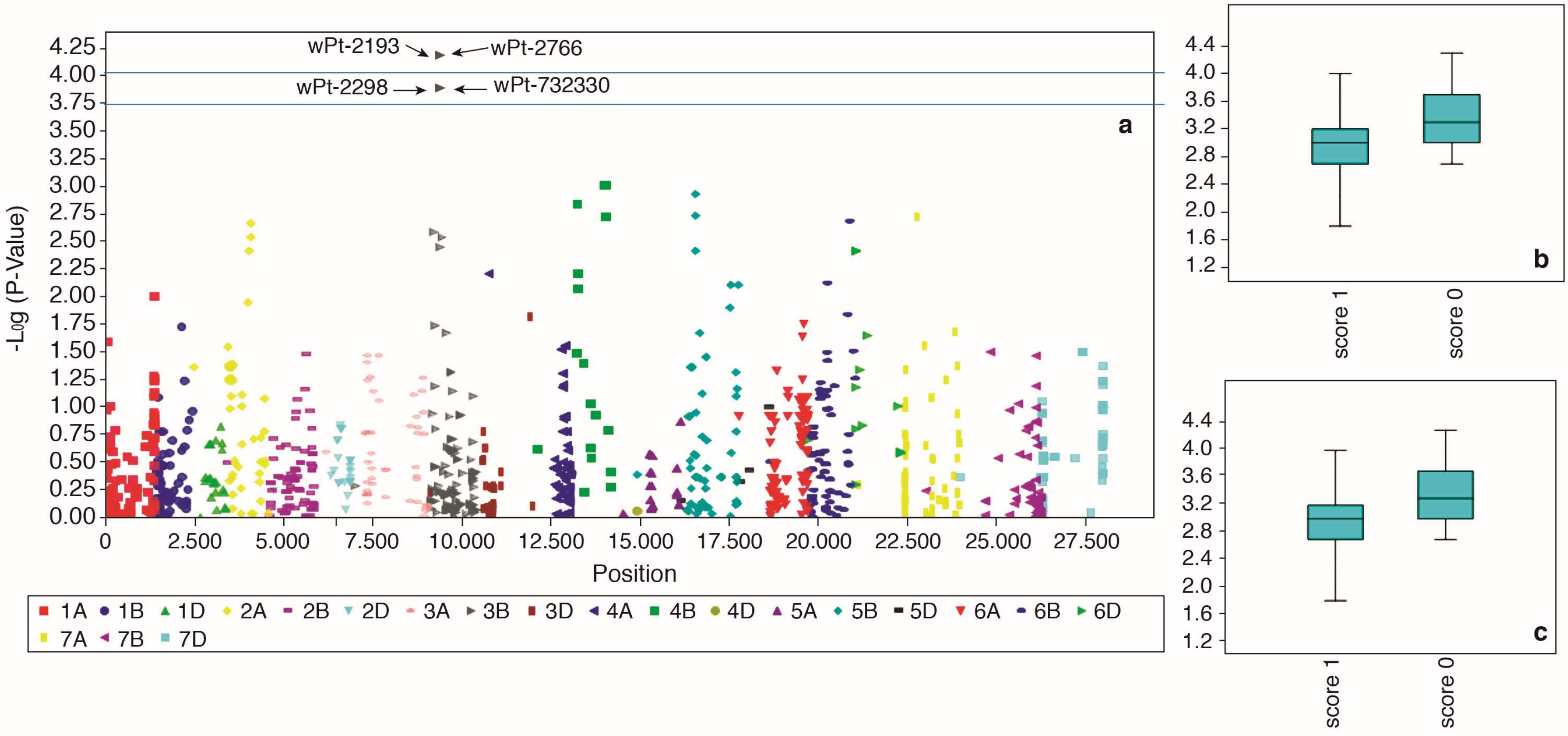
2.5. Marker-Trait Associations for F. culmorum Resistance
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Inoculum Preparation
4.3. Fusarium culmorum Disease Screening
4.3.1. Growth Room
4.3.2. Greenhouse
4.3.3. Field Conditions
4.4. Disease Assessment and Data Analysis
4.5. Genotyping with DArT Markers
4.6. Population Structure Analysis
4.7. Linkage Disequilibrium and GWAS
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Feldman, M. Origin of cultivated wheat. In The World Wheat Book: A History of Wheat Breeding; Bonjean, A.P., Angus, W.J., Eds.; Lavoisier Publishing: Paris, France, 2001; pp. 3–53. [Google Scholar]
- Sohail, Q.; Inoue, T.; Tanaka, H.; Eltayeb, A.E.; Matsuoka, Y.; Tsujimoto, H. Applicability of Aegilops tauschii drought tolerance traits to breeding of hexaploid wheat. Breed. Sci. 2011, 61, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Mujeeb-Kazi, A.; Rosas, V.; Roldan, S. Conservation of the genetic variation of Triticum tauschii in synthetic hexaploid wheats and its potential utilization for wheat improvement. Genet. Resour. Crop Evol. 1996, 43, 129–134. [Google Scholar] [CrossRef]
- Kihara, H. Discovery of the DD-analyser, one of the ancestors of Triticum vulgare. Agric. Hort. 1944, 19, 13–14. [Google Scholar]
- Cox, T.S. Deepening the wheat gene pool. J. Crops Prod. 1998, 1, 1–25. [Google Scholar] [CrossRef]
- Breiman, A.; Graur, D. Wheat Evaluation. Isr. J. Plant Sci. 1995, 43, 58–95. [Google Scholar] [CrossRef]
- Kan, M.; Murat, K.; Mesut, K.; Alexey, M.; Hafi, M.; Fatih, O.; Calvin, Q. Wheat Landraces in Farmers’ Fields in Turkey National Survey, Collection and Conservation, 2009–2014; Food and Agriculture Organization: Quebec City, QC, USA; The United Nations: New York, NY, USA, 2015; 155p. [Google Scholar]
- Chakraborty, S.; Obanor, F.; Westecott, R.; Abeywickrama, K. Wheat crown rot pathogens Fusarium graminearum and F. pseudograminearum lack specialization. Am. Phytopathol. Soc. 2010, 100, 1057–1065. [Google Scholar] [CrossRef] [PubMed]
- Burgess, L.W.; Wearing, A.H.; Toussoun, T.A. Surveys of fusaria associated with crown rot of wheat in Eastern Australia. Aust. J. Agric. Res. 1975, 26, 791–799. [Google Scholar] [CrossRef]
- Tunali, B.; Nicol, J.M.; Hodson, D.; Uckun, Z.; Buyuk, O.; Erdurmus, E.; Hekimhan, H.; Aktas, H.; Akbudak, M.A.; Bagci, S.A. Root and crown rot fungi associated with spring, facultative, and winter wheat in Turkey. Plant Dis. 2008, 98, 1299–1306. [Google Scholar] [CrossRef]
- Saremi, H.; Ammarellou, A.; Jafary, H. Incidence of crown rot disease caused by Fusarium pseudograminearum as a new soil borne fungal species in North West Iran. Pak. J. Biol. Sci. 2007, 27, 3606–3612. [Google Scholar] [CrossRef]
- Hameed, M.A.; Rana, R.M.; Ali, Z. Identification and characterization of a novel Iraqi isolate of Fusarium pseudograminearum causing crown rot in wheat. Genet. Mol. Res. 2012, 11, 1341–1348. [Google Scholar] [CrossRef] [PubMed]
- Paulitz, T.C.; Smiley, R.W.; Cook, R.J. Insights into the prevalence and management of soil borne cereal pathogens under direct seeding in the Pacific Northwest, USA. Can. J. Plant Pathol. 2002, 24, 416–428. [Google Scholar] [CrossRef]
- Smiley, R.W.; Gourlie, J.A.; Easley, S.A. Pathogenicity of fungi associated with wheat crown rot complex in Oregon and Washington. Plant Dis. 2005, 89, 949–957. [Google Scholar] [CrossRef]
- Cook, R.J. Wheat root health management and environmental concern. Can. J. Plant Pathol. 1992, 14, 76–85. [Google Scholar]
- Backhouse, D.; Abubakar, A.A.; Burgess, L.W.; Dennis, J.I.; Hollaway, G.J.; Wildermuth, G.B.; Wallwork, H.; Henry, J.F. Survey of Fusarium species associated with crown rot of wheat and barley in eastern Australia. Aust. Plant Pathol. 2002, 33, 255–261. [Google Scholar] [CrossRef]
- Liu, C.; Ogbonnaya, F.C. Resistance to Fusarium crown rot in wheat and barley: A review. Plant Breed. 2015, 134, 365–372. [Google Scholar] [CrossRef]
- Flint-Garcia, S.A.; Thornsberry, J.M.; Buckler, E.S. Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 2003, 54, 357–374. [Google Scholar] [CrossRef] [PubMed]
- Pariyar, S.R.; Dababat, A.A.; Sannemann, W.; Erginbas-Orakci, G.; Elashry, A.; Siddique, S.; Morgounov, A.; Leon, J.; Grundler, F.M.W. Genome-wide association study in wheat identifies resistance to the cereal cyst nematode Heterodera filipjevi. Phytopathology 2016, 106, 1128–1138. [Google Scholar] [CrossRef] [PubMed]
- Dababat, A.A.; Hugo, G.; Erginbas-Orakci, G.; Susanne Dreisigacker, G.; Imren, M.; Halil Toktay, H.; Elekcioglu, H.; Tesfamariam Mekete, T.; Nicol, J.M.; Omid Ansari, O.; et al. Association analysis of resistance to cereal cyst nematodes (Heterodera avenae) and root lesion nematodes (Pratylenchus neglectus and P. thornei) in CIMMYT advanced spring wheat lines for semi-arid conditions. Breed. Sci. 2016, 66, 692–702. [Google Scholar] [CrossRef] [PubMed]
- Tommasini, L.; Schnurbusch, T.; Fossati, D.; Mascher, F.; Keller, B. Association mapping of Stagonospora nodorum blotch resistance in modern European winter wheat varieties. Theor. Appl. Genet. 2007, 115, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Breseghello, F.; Sorrells, M.E. Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 2006, 172, 1165–1177. [Google Scholar] [CrossRef] [PubMed]
- Neumann, K.; Kobiljski, B.; Denčić, S.; Varshney, R.K.; Börner, A. Genome-wide association mapping: A case study in bread wheat (Triticum aestivum L.). Mol. Breed. 2010. [Google Scholar] [CrossRef]
- Ravel, C.; Praud, S.; Murigneux, A.; Linossier, L.; Dardevet, M.; Balfourier, F.; Dufour, P.; Brunel, D.; Charmet, G. Identification of Glu-B1-1 as a candidate gene for the quantity of high-molecular-weight glutenin in bread wheat (Triticum aestivum L.) by means of an association study. Theor. Appl. Genet. 2006, 112, 738–743. [Google Scholar] [CrossRef] [PubMed]
- Gurung, S.; Mamidi, S.; Bonman, J.M.; Jackson, E.W.; delRıo, L.E.; Acevedo, M.; Mergoum, M.; Adhikari, T.B. Identification of novel genomic regions associated with resistance to Pyrenophoratriticirepentis races 1 and 5 in spring wheat landraces using association analysis. Theor. Appl. Genet. 2011. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.H.; Bai, Y.; Haley, S.D.; Lapitan, N.L.V. Microsatellite—Based molecular diversity of bread wheat germplasm and association mapping of wheat resistance to the Russian wheat aphid. Genetica 2009, 135, 95–122. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Wang, L.; Liu, L.; Zhao, C.; Zheng, Y. Association map—Ping of agronomic traits on chromosome 2A of wheat. Genetica 2009, 137, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Saeed, I.; Chen, X.; Bachir, D.G.; Chen, L.; Hu, Y.G. Association mapping for photosynthesis and yield traits under two moisture conditions and their drought indices in winter bread wheat (Triticum aestivum L.) using SSR markers. Aust. J. Crops Sci. 2017, 11, 248. [Google Scholar] [CrossRef]
- Wallwork, H.; Butt, M.; Cheong, J.P.E.; Williams, K.J. Resistance to crown rot in wheat identified through an improved method for screening adult plants. Aust. Plant Pathol. 2004, 3, 1–7. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Grams, R.A.; Bovill, C.D.; Percy, C.D.; Jolley, R.; Lehmensiek, A.; Wildermuth, G.; Sutherland, M.W. Development of molecular markers for crown rot resistance in wheat: Mapping of QTL for seedling resistance in a ‘2–49’ × ‘Janz’ population. Plant Breed. 2005, 124, 532–537. [Google Scholar] [CrossRef] [Green Version]
- Bovill, W.D.; Horne, M.; Herde, D.; Wildermuth, G.B.; Sutherland, M.W. Pyramiding QTL increases seedling resistance to crown rot (Fusarium pseudograminearum) of wheat (Triticum aestivum). Theor. Appl. Genet. 2010, 121, 127–136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Li, H.; Zhang, C.; Yang, X.; Liu, Y.; Yan, G.; Liu, C. Identification and validation of a major QTL conferring crown rot resistance in hexaploid wheat. Theor. Appl. Genet. 2010, 120, 1119–1128. [Google Scholar] [CrossRef] [PubMed]
- Poole, G.J.; Smiley, R.W.; Paulitz, T.C.; Walker, C.A.; Carter, A.H.; See, D.R.; Garland-Campbell, K. Identification of quantitative trait loci (QTL) for resistance to Fusarium crown rot (Fusarium pseudograminearum) in multiple assay environments in the Pacific Northwestern US. Theor. Appl. Genet. 2012, 125, 91–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.B.; Xie, G.Q.; Ma, J.; Liu, G.R.; Wen, S.M.; Ban, T.; Chakraborty, S.; Liu, C.J. Genetic relationships between resistances to Fusarium head blight and crown rot in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2010, 121, 941–950. [Google Scholar] [CrossRef] [PubMed]
- Castiblanco, V.; Marulanda, J.J.; Würschum, T.; Miedaner, T. Candidate gene based association mapping in Fusarium culmorum for field quantitative pathogenicity and mycotoxin production in wheat. BMC Genet. 2017, 18, 49. [Google Scholar] [CrossRef] [PubMed]
- Sres, A.; Trdan, S.; Leskosek, G.; Vidrih, M.; Vucajnk, F. Effect of spraying speed on head coverage of winter wheat with fungicide, occurrence of Fusarium head blight and deoxynivalenol, yield and grain quality. Cereal Res. Commun. 2015, 43, 72–83. [Google Scholar] [CrossRef] [Green Version]
- Wildermuth, G.B.; McNamara, R.B. Testing wheat seedlings for resistance to crown rot caused by Fusarium graminearum Group 1. Plant Dis. 1994, 78, 949–995. [Google Scholar] [CrossRef]
- Dodman, R.L.; Wildermuth, G.B. Inoculation methods for assessing resistance in wheat to crown rot caused by Fusarium graminearum Group 1. Aust. J. Agric. Res. 1987, 38, 473–486. [Google Scholar]
- Hogg, A.C.; Johnston, R.H.; Dyer, A.T. Applying real-time quantitative PCR to Fusarium crown rot of wheat. Plant Dis. 2007, 91, 1021–1028. [Google Scholar] [CrossRef]
- Mitter, V.; Zhang, M.C.; Liu, C.J.; Ghosh, R.; Ghosh, M.; Chakraborty, S.A. High-throughput bioassay to detect crown rot resistance in wheat germplasm. Plant Pathol. 2006, 55, 433–441. [Google Scholar] [CrossRef]
- Liddell, C.M.; Burgess, L.W.; Taylor, P.W.J. Reproduction of crown rot of wheat caused by Fusarium graminearum group 1 in the greenhouse. Plant Dis. 1986, 70, 632–635. [Google Scholar] [CrossRef]
- Nelson, K.E.; Burgess, L.W. Reaction of Australian cultivars of oats and barley to infection by Fusarium graminearum Group 1. Aust. J. Exp. Agric. 1994, 35, 655–658. [Google Scholar] [CrossRef]
- Erginbas-Orakci, G.; Poole, G.; Nicol, J.M.; Paulitz, T.; Dababat, A.A.; Campbell, K. Assessment of inoculation methods to identify resistance to Fusarium crown rot in wheat. J. Plant Dis. Prot. 2016, 123, 19–27. [Google Scholar] [CrossRef]
- Mujeeb-Kazi, A.; Hettel, G.P. Utilizing Wild Grass Biodiversity in Wheat Improvement: 15 Years of Wide cross Research at CIMMYT; CIMMYT Research Report; No. 2; CIMMYT: Mexico City, Mexico, 1992. [Google Scholar]
- Ogbonnaya, F.C.; Abdalla, O.; Mujeeb-Kazi, A.; Kazi, A.G.; Gosnian, N.; Lagudah, E.S. Synthetic hexaploids: Harnessing species of the primary gene pool for wheat improvement. Plant Breed. Rev. 2013, 37, 35–122. [Google Scholar]
- Van Ginkel, M.; Ogbonnaya, F. Novel genetic diversity from synthetic wheats in breeding cultivars for changing production conditions. Field Crops Res. 2007, 104, 86–94. [Google Scholar] [CrossRef]
- Chao, S.; Dubcovsky, J.; Dvorak, J.; Luo, M.-C.; Baenziger, P.S.; Matnyazov, R.; Clark, D.R.; Talbert, L.E.; Anderson, J.A.; Dreisigacker, S.; et al. Population- and genome-specific patterns of linkage disequilibrium and SNP variation in spring and winter wheat (Triticum aestivum L.). BMC Genom. 2010, 11, 727–744. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, D.; Autrique, E.; Singh, R.; Eillis, M.; Singh, S.; Dreisigacker, S. Identication of genomic regions for grain yield and yield stability and their epistatic interactions. Sci. Rep. 2017, 7, 41578. [Google Scholar] [CrossRef] [PubMed]
- Edae, E.A.; Byrne, P.F.; Haley, S.D.; Lopes, M.S.; Reynolds, M.P. Genome-wide association mapping of yield and yield components of spring wheat under contrasting moisture regimes. Theor. Appl. Genet. 2014, 127, 791–807. [Google Scholar] [CrossRef] [PubMed]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 years of GWAS discovery: Biology, function, and translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef] [PubMed]
- Crossa, J.; Burgueno, J.; Dreisigacker, S.; Vargas, M.; Herrera-Foessel, S.A.; Lillemo, M.; Singh, R.P.; Trethowan, R.; Warburton, M.; Franco, J.; et al. Association analysis of historical bread wheat germplasm using additive genetic covariance of relatives and population structure. Genetics 2007, 177, 1889–1913. [Google Scholar] [CrossRef] [PubMed]
- Somers, D.J.; Banks, T.; DePauw, R.; Fox, S.; Clarke, J.; Pozniak, C.; McCartney, C. Genome-wide linkage disequilibrium analysis in bread wheat and durum wheat. Genome 2007, 50, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Min, D.; Yasir, T.A.; Hu, Y.G. Genetic diversity, population structure and linkage disequilibrium in elite Chinese winter wheat investigated with SSR markers. PLoS ONE 2012, 7, e44510. [Google Scholar] [CrossRef] [PubMed]
- Chao, S.; Zhang, W.; Dubcovsky, J.; Sorrells, M. Evaluation of genetic diversity and genome-wide linkage disequilibrium among US wheat (Triticum aestivum L.) germplasm representing different market class. Crop Sci. 2007, 47, 1018–1030. [Google Scholar] [CrossRef]
- Benson, J.; Brown-Guedira, G.; Murphy, J.P.; Sneller, C. Population structure, linkage disequilibrium, and genetic diversity in soft winter wheat enriched for fusarium head blight resistance. Plant Genome 2012, 5, 71–80. [Google Scholar] [CrossRef]
- Zhang, D.D.; Bai, G.H.; Zhu, C.S.; Yu, J.M.; Carver, B.F. Genetic Diversity, Population Structure and Linkage Disequilibrium in U.S. Elite Winter Wheat. Plant Genome 2010, 3, 117–127. [Google Scholar] [CrossRef]
- Würschum, T.; Langer, S.M.; Longin, C.F.; Korzun, V.; Akhunov, E.; Ebmeyer, E.; Schachschneider, R.; Schacht, J.; Kazman, E.; Reif, J.C. Population structure, genetic diversity and linkage disequilibrium in elite winter wheat assessed with SNP and SSR markers. Theor. Appl. Genet. 2013, 126, 1477–1486. [Google Scholar] [CrossRef] [PubMed]
- Mora, F.; Castillo, D.; Lado, B.; Matus, I.; Poland, J.; Belzile, F.; von Zitzewitz, J.; del Pozo, A. Genome-wide association mapping of agronomic traits and carbon isotope discrimination in a worldwide germplasm collection of spring wheat using SNP markers. Mol. Breed. 2015, 35, 69. [Google Scholar] [CrossRef]
- Massman, J.; Cooper, B.; Horsley, R.; Neate, S.; Dill-Macky, R.; Chao, S.; Dong, Y.; Schwarz, P.; Muehlbauer, G.J.; Smith, K.P. Genome-wide association mapping of Fusarium head blight resistance in contemporary barley breeding germplasm. Mol. Breed. 2011, 27, 439–454. [Google Scholar] [CrossRef]
- Yu, L.X.; Morgounov, A.; Wanyera, R.; Keser, M.; Singh, S.K.; Sorrells, M.E. Identification of Ug99 stem rust resistance loci in winter wheat germplasm using genome-wide association analysis. Theor. Appl. Genet. 2012, 125, 749–758. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Dell, B.; Biddulph, B.; Drake-Brockman, F.; Walker, E.; Khan, N.; Wong, D.; Hayden, M.; Appels, R. Wild-type alleles of Rht-B1and Rht-D1 as independent determinants of thousand-grain weight and kernel number per spike in wheat. Mol. Breed. 2013, 32, 771–783. [Google Scholar] [CrossRef]
- Rasheed, A.; Xia, X.; Ogbonnaya, F.; Mahmood, T.; Zongwen, Z.; Mujeeb-Kazi, A.; He, Z. Genome-wide association for grain morphology in synthetic hexaploid wheats using digital imaging analysis. BMC Plant Biol. 2014, 14, 128. [Google Scholar] [CrossRef] [PubMed]
- Maccaferri, M.; Zhang, J.; Bulli, P.; Abate, Z.; Chao, S.; Cantu, D.; Bossolini, E.; Chen, X.; Pumphrey, M.; Dubcovsky, J.A. Genome-wide association study of resistance to stripe rust (Pucciniastriiformis f. sp. tritici) in a worldwide collection of hexaploid spring wheat (Triticum aestivum L.). G3 2015, 5, 449–465. [Google Scholar] [CrossRef] [PubMed]
- Hong, E.; Park, J. Sample size and statistical power calculations. Genom. Inform. 2012, 10, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Bovill, W.D.; Ma, W.; Ritter, K.; Collard, B.C.Y.; Davis, M.; Wildermuth, G.B.; Sutherland, M.W. Identification of novel QTL for resistance to crown rot in the doubled haploid wheat population ‘W21MMT70’ × ‘Mendos’. Plant Breed. 2006, 125, 538–543. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Z.; Kilian, A.; Yan, G.; Liu, C.J. QTL Conferring Fusarium Crown Rot Resistance in the Elite Bread Wheat Variety EGA Wylie. PLoS ONE 2014, 9, e96011. [Google Scholar] [CrossRef] [PubMed]
- Zadoks, J.C.; Chang, T.T.; Konsak, C.F.A. A decimal code for the growth stages of cereals. Eucarpia Bull. 1974, 7, 11. [Google Scholar] [CrossRef]
- Saghai-Maroof, M.A.; Soliman, K.; Jorgensen, R.A.; Allard, R.W. Ribosomal DNA spacer length polymorphism in barley: Mendelian inheritance, chromosomal location and population dynamics. Proc. Natl. Acad. Sci. USA 1984, 81, 8014–8018. [Google Scholar] [CrossRef] [PubMed]
- Dreisigacker, S.; Sehgal, D.; Reyes Jaimez, A.E.; Luna Garrido, B.; Muñoz Zavala, S.; Núñez Ríos, C.; Mollins, J.; Mall, S. CIMMYT Wheat Molecular Genetics: Laboratory Protocols and Applications to Wheat Breeding; CIMMYT: Mexico City, Mexico, 2016. [Google Scholar]
- Wenzel, P.; Carling, J.; Kudrna, D.; Jaccoud, D.; Huttner, E. Diversity Arrays Technology (DArT) for Whole-Genome Profiling of Barley; Ronald, M., Phillips, L., Eds.; Triticarte Pty. Ltd.: St. Paul, MN, USA; Canberra, Australia, 2004. [Google Scholar]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [PubMed]
- Pritchard, J.K.; Stephens, M.; Rosenberg, N.A.; Donnelly, P. Association Mapping in Structured Populations. Am. J. Hum. Genet. 2000, 67, 170–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Falush, D.M.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics 2003, 164, 1567–1587. [Google Scholar] [PubMed]
- Bradbury, P.J.; Zhang, Z.; Kroon, D.E.; Casstevens, T.M.; Ramdoss, Y.; Buckler, E.S. TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 2003, 23, 2633–2635. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.E.; George, A.W.; Forrest, K.L.; Kilian, A.; Hayden, M.J.; Morell, M.K.; Cavanagh, C.R. A multiparent advanced generation inter-cross population for genetic analysis in wheat. Plant Biotechnol. J. 2002, 9, 826–839. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar]




| Cross Name | CID | Adult-Plant Resistance | Seedling Resistance | |||
|---|---|---|---|---|---|---|
| GHSS | GHCS | FCS | GRCS | RR | ||
| BABAX/LR42//BABAX/3/BABAX/LR42//BABAX/4/T.DICOCCON PI94625/AE.SQUARROSA (372)//3*PASTOR/5/T.DICOCCON PI94625/AE.SQUARROSA (372)//3*PASTOR | 6,000,229 | 2.2 | 2.5 | 2.2 | 2.5 | A-S |
| BABAX/LR42//BABAX/3/BABAX/LR42//BABAX/4/T.DICOCCON PI94625/AE.SQUARROSA (372)//3*PASTOR/5/T.DICOCCON PI94625/AE.SQUARROSA (372)//3*PASTOR | 6,000,238 | 1.5 | 2.2 | 2.7 | 2.2 | S |
| FRET2*2/4/SNI/TRAP#1/3/KAUZ*2/TRAP//KAUZ/5/ONIX | 6,000,365 | 1.5 | 2.5 | 3 | 1.8 | S |
| ACHTAR/4/MILAN/KAUZ//PRINIA/3/BAV92 | 6,000,537 | 2.2 | 2.5 | 2.5 | 2.3 | A-S |
| SOKOLL*2/ROLF07 | 6,000,973 | 1.5 | 2.5 | 2.3 | 3.2 | A |
| GK ARON/AG SECO 7846//2180/4/2*MILAN/KAUZ//PRINIA/3/BAV92 | 6,001,016 | 2.5 | 3.5 | 3.2 | 1.8 | S |
| SOKOLL//FRTL/2*PIFED | 6,001,172 | 2.2 | 2.8 | 2.5 | 2.3 | A-S |
| ROLF07/3/T.DICOCCON PI94625/AE.SQUARROSA (372)//3*PASTOR | 6,001,240 | 2.2 | 2.8 | 3 | 2.3 | S |
| CUNNINGHAM/4/SNI/TRAP#1/3/KAUZ*2/TRAP//KAUZ | 6,001,457 | 2.2 | 2.8 | 3 | 1.8 | S |
| SOKOLL*2/4/CHEN/AEGILOPS SQUARROSA (TAUS)//FCT/3/STAR | 6,001,643 | 2.5 | 3.5 | 2.7 | 2.3 | S |
| SERI*3//RL6010/4*YR/3/PASTOR/4/BAV92/5/MONARCA F2007/6/PVN//CAR422/ANA/5/BOW/CROW//BUC/PVN/3/YR/4/TRAP#1 | 6,000,104 | 2.2 | 2.3 | 2.5 | 2.8 | A |
| CNO79//PF70354/MUS/3/PASTOR/4/BAV92/5/FRET2/KUKUNA//FRET2/6/MILAN/KAUZ//PRINIA/3/BAV92 | 6,000,615 | 1.5 | 2.8 | 3.5 | 2.3 | S |
| 2–49 | 1.3 | 2.3 | 2.5 | 2.5 | MR | |
| Altay | 1.2 | 2.5 | 2.3 | 2.4 | MR | |
| Sunco | 1.5 | 2.5 | 2.3 | 2.6 | MR | |
| Seri | 2.2 | 3.2 | 3.5 | 4 | MS | |
| Kiziltan | 2.5 | 3.5 | 4 | 4 | HS | |
| Kutluk | 2.5 | 3.5 | 4 | 4 | HS | |
| Wylie | 2.3 | 3.3 | 3.4 | 3.5 | S | |
| Janz | 2.5 | 3.5 | 3.7 | 4 | HS | |
| LSD | 0.5 | 0.53 | 1.04 | 1.08 | ||
| Probability | <0.001 | <0.001 | <0.001 | <0.001 | ||
| GHCS | FCR | GRCS | GHSS | |
|---|---|---|---|---|
| GHCS | 1.00000 | 0.10153 | 0.01128 | 0.50728 |
| FCR | 0.2388 | 1.00000 | 0.31369 | 0.14685 |
| GRCS | 0.89671 | 0.00010 | 1.00000 | 0.13237 |
| GHSS | 0.00000 | 0.08574 | 0.12258 | 1.00000 |
| Trait | Marker | CHR | POS (cM) | p-Value | FDR Adjusted p-Values | r2 (%) |
|---|---|---|---|---|---|---|
| FCS (GLM) | wPt-2193 | 3B | 39.1 | 6.56 × 10−5 | 0.01232 | 11.4 |
| FCS (GLM) | wPt-2766 | 3B | 39.1 | 6.56 × 10−5 | 0.01232 | 11.4 |
| GHSS (MLM) | wPt-669517 | 2D | 95.7 | 3.24 × 10−5 | 0.01997 | 11.6 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erginbas-Orakci, G.; Sehgal, D.; Sohail, Q.; Ogbonnaya, F.; Dreisigacker, S.; Pariyar, S.R.; Dababat, A.A. Identification of Novel Quantitative Trait Loci Linked to Crown Rot Resistance in Spring Wheat. Int. J. Mol. Sci. 2018, 19, 2666. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19092666
Erginbas-Orakci G, Sehgal D, Sohail Q, Ogbonnaya F, Dreisigacker S, Pariyar SR, Dababat AA. Identification of Novel Quantitative Trait Loci Linked to Crown Rot Resistance in Spring Wheat. International Journal of Molecular Sciences. 2018; 19(9):2666. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19092666
Chicago/Turabian StyleErginbas-Orakci, Gul, Deepmala Sehgal, Quahir Sohail, Francis Ogbonnaya, Susanne Dreisigacker, Shree R. Pariyar, and Abdelfattah A. Dababat. 2018. "Identification of Novel Quantitative Trait Loci Linked to Crown Rot Resistance in Spring Wheat" International Journal of Molecular Sciences 19, no. 9: 2666. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19092666





