Genome-Wide Association Studies of 39 Seed Yield-Related Traits in Sesame (Sesamum indicum L.)
Abstract
:1. Introduction
2. Results
2.1. Variability and Correlation of the Seed Yield-Related Traits in the Sesame Association Panel
2.2. Genetic Variants Associated with Seed Yield-Related Traits in Sesame
2.3. Comparing Previous QTLs on Seed Yield-Related Traits from Bi-Parental Linkage Mapping with Our GWAS Results
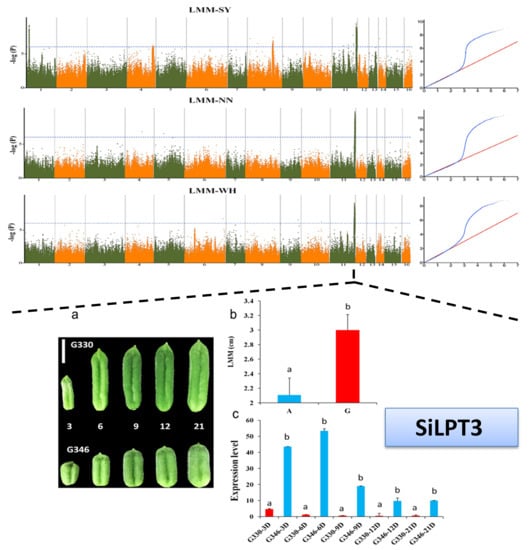
2.4. Important Candidate Genes Associated with Seed Yield in Sesame
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Field Growth Conditions
4.3. Trait Evaluation
4.4. Statistical Analysis
4.5. Genome Wide Association Study Implementation
4.6. Candidate Gene Mining
4.7. Gene Expression Analysis Based on Quantitative Real-Time PCR
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CN | effective capsule number in plant |
| CNB | effective capsule number in branch |
| CNM | effective capsule number in main stem |
| CPLM | dry capsule pericarp weight of lateral capsule in main stem |
| CPMB | dry capsule pericarp weight of medial capsule in branch |
| CPMM | dry capsule pericarp weight of medial capsule in main stem |
| DNA | deoxyribonucleic acid |
| GWAS | genome wide association study |
| LCNB | effective lateral capsule number in branch |
| LCNM | effective lateral capsule number in main stem |
| LD | linkage disequilibrium |
| LG | linkage group |
| LLB | length of lateral capsule in branch |
| LLM | length of lateral capsule in main stem |
| LMB | length of medial capsule in branch |
| LMM | length of medial capsule in main stem |
| MAF | minor allele frequency |
| MCNB | effective medial capsule number in branch |
| MCNM | effective medial capsule number in main stem |
| NN | Nanning |
| qRT-PCR | quantitative real-time polymerase chain reaction |
| QTL | quantitative trait loci |
| RLM | ratio of seed weight and capsule pericarp weight for lateral capsule in main stem |
| RMB | ratio of seed weight and capsule pericarp weight for medial capsule in branch |
| RMM | ratio of seed weight and capsule pericarp weight for medial capsule in main stem |
| RNA | ribonucleic acid |
| SNLB | seed number per lateral capsule in branch |
| SNLM | seed number per lateral capsule in main stem |
| SNMB | seed number per medial capsule in branch |
| SNMM | seed number per medial capsule in main stem |
| SNP | single nucleotide polymorphism |
| SY | Sanya |
| SWLB | dry seed weight of per lateral capsule in branch |
| SWLM | dry seed weight of per lateral capsule in main stem |
| SWMB | dry seed weight of per medial capsule in branch |
| SWMM | dry seed weight of per medial capsule in main stem |
| TLM | thickness of lateral capsule in main stem |
| TMB | thickness of medial capsule in branch |
| TMM | thickness of medial capsule in main stem |
| TSW | thousand seeds weight |
| WH | Wuhan |
| WLM | width of lateral capsule in main stem |
| WMB | width of medial capsule in branch |
| WMM | width of medial capsule in main stem |
| YB | yield of branch per plant |
| YLB | yield of lateral capsules in branch |
| YLM | yield of lateral capsules in main stem |
| YM | yield of main stem per plant |
| YMB | yield of medial capsules in branch |
| YMM | yield of medial capsules in main stem |
| YP | yield per plant |
References
- Anilakumar, K.R.; Pal, A.; Khanum, F.; Bawa, A.S. Nutritional, medicinal and industrial uses of sesame (Sesamum indicum L.) seeds: An overview. Agric. Conspec. Sci. 2010, 75, 159–168. [Google Scholar]
- Dossa, K.; Wei, X.; Niang, M.; Liu, P.; Zhang, Y.; Wang, L.; Liao, B.; Cissé, N.; Zhang, X.; Diouf, D. Near-infrared reflectance spectroscopy reveals wide variation in major components of sesame seeds from Africa and Asia. Crop J. 2018, 6, 202–206. [Google Scholar] [CrossRef]
- Ingersent, K.A. World agriculture: Towards 2015/2030–An FAO perspective. J. Agric. Econ. 2003, 54, 513–515. [Google Scholar]
- Langham, D.R. Phenology of Sesame. In Issues in New Crops and New Uses; Janick, J., Whipley, A., Eds.; ASHS Press: Alexandria, VA, USA, 2007; p. 39. [Google Scholar]
- Dossa, K.; Diouf, D.; Wang, L.; Wei, X.; Zhang, Y.; Niang, M.; Fonceka, D.; Yu, J.; Mmadi, M.A.; Yehouessi, L.W.; et al. The emerging oilseed crop Sesamum indicum enters the “Omics” era. Front. Plant Sci. 2017, 8, 1154. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organization Statistical Databases (FAOSTAT). 2017. Available online: http://faostat.fao.org/ (accessed on 19 March 2018).
- Akhtar, K.P.; Sarwar, G.; Dickinson, M.; Ahmad, M.; Haq, M.A.; Hameed, S.; Iqbal, M.J. Sesame phyllody disease: Its symptomatology, etiology, and transmission in Pakistan. Turk. J. Agric. For. 2009, 33, 477–486. [Google Scholar]
- Yol, E.; Uzun, B. Geographical patterns of sesame accessions grown under Mediterranean environmental conditions, and establishment of a core collection. Crop Sci. 2012, 52, 2206–2214. [Google Scholar] [CrossRef]
- Biabani, A.R.; Pakniyat, H. Evaluation of seed yield-related characters in sesame (Sesamum indicum L.) using factor and path analysis. Pak. J. Biol. Sci. 2008, 11, 1157–1160. [Google Scholar] [PubMed]
- Wu, K.; Liu, H.; Yang, M.; Tao, Y.; Ma, H.; Wu, W.; Zuo, Y.; Zhao, Y. High-density genetic map construction and QTLs analysis of grain yield-related traits in sesame (Sesamum indicum L.) based on RAD-Seq technology. BMC Plant. Biol. 2014, 14, 274. [Google Scholar] [CrossRef] [PubMed]
- Diouf, M.; Boureima, S.; Diop, T.; Çagirgan, M. Gamma rays-induced mutant spectrum and frequency in sesame. Turk. J. Field Crops 2010, 15, 99–105. [Google Scholar]
- Mosjidis, J.A.; Yermanos, D.M. Plant position effect on seed weight, oil content, and oil composition in sesame. Euphytica 1985, 34, 193–199. [Google Scholar] [CrossRef]
- Tashiro, T.; Fukuda, Y.; Osawa, T. Oil contents of seeds and minor components in the oil of sesame, Sesamum indicum L.; as affected by capsule position. Jpn. J. Crop Sci. 1991, 60, 116–121. [Google Scholar] [CrossRef]
- Kumazaki, T.; Yamada, Y.; Karaya, S.; Kawamura, M.; Hirano, T.; Yasumoto, S.; Katsuta, M.; Michiyama, H. Effects of day length and air and soil temperatures on sesamin and sesamolin contents of sesame seed. Plant Prod. Sci. 2009, 12, 481–491. [Google Scholar] [CrossRef]
- Huang, X.; Han, B. Natural variations and genome-wide association studies in crop plants. Annu. Rev. Plant Biol. 2014, 65, 531–551. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Liu, K.; Zhang, Y.; Feng, Q.; Wang, L.; Zhao, Y.; Li, D.; Zhao, Q.; Zhu, X.; Zhu, X.; et al. Genetic discovery for oil production and quality in sesame. Nat. Commun. 2015, 6, 8609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Dossa, K.; Zhang, Y.; Wei, X.; Wang, L.; Zhang, Y.; Liu, A.; Zhou, R.; Zhang, X. GWAS uncovers differential genetic bases for drought and salt tolerances in sesame at the germination stage. Genes 2018, 9, 87. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yu, S.; Tong, C.; Zhao, Y.; Liu, Y.; Song, C.; Zhang, Y.; Zhang, X.; Wang, Y.; Hua, W.; et al. Genome sequencing of the high oil crop sesame provides insight into oil biosynthesis. Genome Biol. 2014, 15, R39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dossa, K. A physical map of important QTLs, functional markers and genes available for sesame breeding programs. Physiol. Mol. Biol. Plants 2016, 22, 613–619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.; Suh, S.S.; Park, E.; Cho, E.; Ahn, J.H.; Kim, S.G.; Lee, J.S.; Kwon, Y.M.; Lee, I. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Gene Dev. 2000, 14, 2366–2376. [Google Scholar] [CrossRef] [PubMed]
- Tsuchisaka, A.; Theologis, A. Heterodimeric interactions among the 1-amino-cyclopropane-1-carboxylate synthase polypeptides encoded by the Arabidopsis gene family. Proc. Natl. Acad. Sci. USA 2004, 101, 2275–2280. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.X.; Malitsky, S.; De Oliveira, S.; Branigan, C.; Franke, R.B.; Schreiber, L.; Aharoni, A. SHINE transcription factors act redundantly to pattern the archetypal surface of Arabidopsis flower organs. PLoS Genet. 2011, 7, e1001388. [Google Scholar] [CrossRef] [PubMed]
- Klepikova, A.V.; Kasianov, A.S.; Gerasimov, E.S.; Logacheva, M.D.; Penin, A.A. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 2016, 88, 1058–1070. [Google Scholar] [CrossRef] [PubMed]
- Rao, P.V.R.; Prasuna, K.; Anuradha, G.; Srividya, A.; Vemireddy, L.R.; Shankar, V.G.; Sridhar, S.; Jayaprada, M.; Reddy, K.R.; Reddy, N.E.; et al. Molecular mapping of important agro-botanic traits in sesame. Electron. J. Plant Breed. 2014, 5, 475–488. [Google Scholar]
- Shim, K.B.; Shin, S.H.; Shon, J.Y.; Kang, S.G.; Yang, W.H.; Heu, S.G. Classification of a collection of sesame germplasm using multivariate analysis. J. Crop Sci. Biotechnol. 2006, 19, 151–155. [Google Scholar] [CrossRef]
- Monpara, B.A.; Khairnar, S.S. Heritability and expected genetic gain from selection in components of crop duration and seed yield in sesame (Sesamum indicum L.). Plant Gene and Trait 2016, 7, 1–5. [Google Scholar]
- Emamgholizadeh, S.; Parsaeian, M.; Baradaran, M. Seed yield prediction of sesame using artificial neural network. Eur. J. Agron. 2015, 68, 89–96. [Google Scholar] [CrossRef]
- Ramazani, S.H.R. Surveying the relations among traits affecting seed yield in sesame (Sesamum indicum L.). J. Crop Sci. Biotechnol. 2016, 19, 303–309. [Google Scholar] [CrossRef]
- Boyles, R.E.; Cooper, E.A.; Myers, M.T.; Brenton, Z.; Rauh, B.L.; Morris, G.P.; Kresovich, S. Genome-wide association studies of grain yield components in diverse sorghum germplasm. Plant Genome 2016, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Korte, A.; Farlow, A. The advantages and limitations of trait analysis with GWAS: A review. Plant Meth. 2013, 9, 29. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Peng, L.; Zhang, C.; Lu, J.; Yang, B.; Xiao, Z.; Liang, Y.; Xu, X.; Qu, C.; Zhang, K.; et al. Genome-wide association and transcriptome analyses reveal candidate genes underlying yield-determining traits in Brassica napus. Front. Plant Sci. 2017, 8, 206. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Wang, H.; Liu, S.; Li, Z.; Yang, X.; Yan, J.; Li, J.; Tran, L.S.P.; Qin, F. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat. Com. 2015, 6, 8326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, Z.; Wang, X.; Liu, Z.; Gu, Q.; Zhang, Y.; Li, Z.; Ke, H.; Yang, J.; Wu, J.; Wu, L.; et al. Genome-wide association study discovered genetic variation and candidate genes of fibre quality traits in Gossypium hirsutum L. Plant Biotechnol. J. 2017, 15, 982–996. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.; Basu, A.; Kundu, S. Overexpression of a new osmotin- like protein gene (SindOLP) confers tolerance against biotic and abiotic stresses in sesame. Front. Plant Sci. 2017, 8, 410. [Google Scholar] [CrossRef] [PubMed]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008; ISBN 3-900051-07-0. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar]
- Wei, T.; Simko, V. Corrplot: Visualization of a Correlation Matrix, R Package Version 0.77. 2016. Available online: http://CRAN.R-project.org/package=corrplot (accessed on 15 March 2018).
- Kang, H.; Sul, J.; Service, S.; Zaitlen, N.; Kong, S.; Freimer, N.; Sabatti, C.; Eskin, E. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 2010, 42, 348–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.X.; Yeung, J.M.; Cherny, S.S.; Sham, P.C. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum. Genet. 2012, 131, 747–756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar]
- Conesa, A.; Götz, S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genom. 2008, 2008, 619832. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Fang, L.; Zheng, H.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L.; et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Mmadi, M.A.; Dossa, K.; Wang, L.; Zhou, R.; Wang, Y.; Cisse, N.; Sy, M.O.; Zhang, X. Functional characterization of the versatile MYB gene family uncovered their important roles in plant development and responses to drought and waterlogging in sesame. Genes 2017, 8, 362. [Google Scholar] [CrossRef] [PubMed]
- Lalitha, S. Primer premier 5. Biotechnol. Softw. Int. Rep. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]








| LG | Position (bp) | Env. | Traits | PVE (%) | LG | Position (bp) | Env. | Traits | PVE (%) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 1,700,170 | SY | CNB | 7.00 | 5 | 17,411,684 | SY | SNMM | 7.29 |
| CN | 7.10 | SWMM | 6.79 | ||||||
| 1 | 1,994,183 | SY | YB | 8.68 | 6 | 3,404,764 | SY | YB | 10.65 |
| YMB | 7.41 | MCNB | 7.33 | ||||||
| 1 | 4,450,107 | SY | YB | 7.55 | YMB | 11.15 | |||
| YMB | 6.99 | 6 | 3,790,583 | SY | TWB | 7.13 | |||
| 1 | 6,149,415 | SY | TMM | 7.61 | WMB | 8.28 | |||
| WMM | 6.58 | 6 | 5,995,560 | SY | YB | 10.91 | |||
| 1 | 8,185,969 | SY | YB | 7.03 | CNB | 6.99 | |||
| YMB | 7.26 | YMB | 11.32 | ||||||
| 1 | 9,906,190 | SY | YB | 6.67 | 6 | 9,021,538 | SY | YB | 8.12 |
| YMB | 6.88 | YMB | 8.34 | ||||||
| 1 | 11,118,941 | SY | SNMB | 6.32 | 6 | 9,971,772 | NN | TMM | 6.29 |
| SWMB | 6.50 | WMM | 6.39 | ||||||
| 1 | 17,291,730 | SY | YB | 8.73 | 6 | 14,154,329 | SY | YB | 7.29 |
| YMB | 9.10 | YMB | 6.59 | ||||||
| 2 | 1,201,448 | SY | YB | 8.76 | 6 | 14,581,641 | SY | LCNM | 10.29 |
| YMB | 9.15 | CNM | 6.41 | ||||||
| YB | 6.41 | 6 | 14,701,957 | WH | TMM | 7.09 | |||
| 2 | 5,260,400 | SY | YB | 7.08 | WMM | 6.96 | |||
| YMB | 7.87 | 6 | 15,551,496 | SY | RMM | 7.63 | |||
| 2 | 6,057,670 | NN | TMM | 9.46 | SNMM | 6.13 | |||
| WMM | 11.33 | 6 | 21,992,131 | SY | YB | 10.89 | |||
| 2 | 7,236,995 | SY | YB | 6.09 | YMB | 10.77 | |||
| YMB | 6.11 | 7 | 6,763,527 | SY | RMB | 6.69 | |||
| 2 | 8,388,879 | NN | TWB | 6.32 | SWMB | 7.65 | |||
| TWB | 7.24 | 7 | 1,702,826 | NN | CNM | 9.42 | |||
| 2 | 9,244,103 | SY | TMM | 6.04 | WH | CNM | 6.66 | ||
| WH | WMM | 9.85 | 8 | 1,398,196 | SY | YB | 11.12 | ||
| TMM | 9.25 | YMB | 11.99 | ||||||
| 2 | 11,245,765 | SY | TMM | 7.95 | 8 | 1,668,572 | SY | YB | 7.38 |
| WMM | 7.56 | SNMM | 9.89 | ||||||
| 2 | 15,016,082 | SY | YB | 7.51 | YM | 6.66 | |||
| YMB | 7.78 | SWMM | 8.84 | ||||||
| 2 | 17,451,873 | SY | SNMB | 6.47 | 8 | 21,325,953 | SY | YB | 8.97 |
| RMM | 9.17 | YMM | 7.68 | ||||||
| SWMM | 14.89 | 9 | 1,007,867 | SY | RMM | 8.57 | |||
| SWMB | 6.83 | SNMM | 8.58 | ||||||
| SNMM | 15.13 | 9 | 954,526 | WH | SNMM | 7.48 | |||
| 3 | 4,840,197 | SY | LCNM | 6.12 | WMM | 8.03 | |||
| CNM | 6.42 | TMM | 7.80 | ||||||
| 3 | 13,198,513 | SY | YB | 6.68 | 10 | 1,647,805 | SY | CNB | 12.64 |
| YMB | 7.24 | CN | 6.14 | ||||||
| 3 | 14,990,430 | SY | YB | 8.68 | 10 | 3,823,922 | SY | CNB | 7.43 |
| CNB | 9.34 | MCNB | 6.85 | ||||||
| MCNB | 9.40 | 10 | 7,418,158 | NN | TMM | 6.24 | |||
| YMB | 8.81 | WMM | 6.31 | ||||||
| 3 | 16,939,689 | SY | YB | 6.70 | 10 | 8,305,398 | SY | YB | 7.76 |
| YMB | 6.63 | YMB | 7.89 | ||||||
| 3 | 20,410,997 | SY | TMM | 6.47 | 10 | 10,792,029 | SY | YB | 7.29 |
| WMM | 6.79 | YMB | 6.60 | ||||||
| 3 | 20,876,555 | SY | YB | 10.76 | 10 | 12,008,065 | SY | RMM | 6.83 |
| YMB | 11.09 | SNMM | 6.61 | ||||||
| 3 | 20,878,243 | SY | CNB | 8.06 | 10 | 14,650,964 | SY | YB | 6.81 |
| MCNB | 8.44 | YMM | 7.50 | ||||||
| 3 | 24,164,350 | SY | TMM | 6.84 | 10 | 15,097,365 | SY | TMM | 7.41 |
| WMM | 6.43 | WMM | 6.90 | ||||||
| 4 | 2,505,014 | SY | YB | 7.98 | 11 | 6,996,833 | SY | SNMM | 6.53 |
| YMB | 7.82 | SWMM | 7.78 | ||||||
| 4 | 6,419,408 | SY | WMM | 6.99 | 11 | 11,923,935 | SY | YB | 7.10 |
| CPMM | 6.25 | YMB | 7.77 | ||||||
| 4 | 14,211,075 | SY | YB | 7.47 | 11 | 14,876,966 | SY | YB | 6.33 |
| YMB | 6.10 | YMB | 6.91 | ||||||
| 5 | 202,984 | SY | YB | 9.50 | 11 | 15,137,600 | SY | TMM | 7.52 |
| YMM | 6.92 | WMM | 6.55 | ||||||
| 5 | 2,854,336 | NN | YM | 7.85 | 12 | 328,609 | SY | YM | 7.53 |
| CNM | 8.20 | YMM | 7.78 | ||||||
| 5 | 5,479,753 | SY | YB | 7.61 | 12 | 2,356,955 | SY | YB | 6.24 |
| CNB | 8.89 | YMB | 6.72 | ||||||
| YMB | 7.81 | 12 | 4,200,237 | SY | YB | 6.60 | |||
| CNM | 13.68 | YMB | 7.21 | ||||||
| NN | CNM | 17.90 | 12 | 4,895,688 | SY | SNMM | 8.67 | ||
| 5 | 6,737,753 | SY | CNB | 8.89 | SWMM | 6.18 | |||
| CNM | 13.68 | 13 | 2,772,629 | SY | YB | 8.17 | |||
| NN | CNM | 17.90 | YMB | 8.49 | |||||
| 5 | 6,738,735 | NN | LCNM | 7.01 | 14 | 194,410 | SY | YB | 6.20 |
| YMB | 6.70 | ||||||||
| CNM | 13.30 | 15 | 2,174,040 | SY | YB | 7.20 | |||
| SY | LCNM | 13.03 | YMB | 7.22 | |||||
| WH | CNM | 14.79 | 15 | 2,372,143 | WH | YB | 6.29 | ||
| 5 | 6,757,688 | NN | SNMB | 8.02 | CNM | 7.03 | |||
| SWMB | 10.88 | YM | 6.01 | ||||||
| 5 | 9,869,746 | NN | RMB | 6.54 | 15 | 3,989,016 | SY | YB | 6.72 |
| SWMB | 6.08 | YMB | 6.69 | ||||||
| 5 | 11,806,702 | SY | TWB | 6.29 | 16 | 555,771 | WH | TMM | 7.55 |
| TWB | 6.34 | WMM | 6.40 | ||||||
| 5 | 15,855,382 | WH | SNMM | 6.76 | 16 | 1,633,469 | SY | YB | 7.97 |
| TMM | 9.11 | YMB | 8.17 | ||||||
| WMM | 8.68 | 16 | 2,989,809 | SY | CNB | 9.26 | |||
| NN | WMM | 6.23 | MCNB | 8.26 | |||||
| 5 | 17,340,920 | SY | CNB | 8.64 | |||||
| MCNB | 10.45 | ||||||||
| Traits Linkage Mapping | Code | LG | Start (bp) | End (bp) | Traits GWAS | LG | SNP Position (bp) |
|---|---|---|---|---|---|---|---|
| Grain number per capsule | Qgn-6 | 6 | 1,739,987 | 2,125,872 | YB | 6 | 1,741,236 |
| YLB | 6 | 2,081,828 | |||||
| Capsule number per plant | Qcn-11 | 9 | 6,032,193 | 8,312,219 | MCNM | 9 | 5,988,865 |
| CNB | 9 | 7,589,997 | |||||
| MCNB | 9 | 7,839,050 | |||||
| Capsule length | Qcl-3 | 3 | 1,566,853 | 2,593,783 | YB | 3 | 2,588,239 |
| YMB | 3 | 2,588,241 | |||||
| Qcl-4 | 5 | 9,840,981 | 10,961,395 | YLB | 5 | 9,857,730 | |
| RMB | 5 | 9,869,746 | |||||
| SWMB | 5 | 9,869,746 | |||||
| TMM | 5 | 9,895,178 | |||||
| WMB | 5 | 9,974,401 | |||||
| TMB | 5 | 10,197,769 | |||||
| TMB | 5 | 10,208,013 | |||||
| YLB | 5 | 10,705,889 | |||||
| SWMB | 5 | 10,773,145 | |||||
| WMB | 5 | 10,781,532 | |||||
| LLM | 5 | 10,786,506 | |||||
| CN | 5 | 10,786,597 | |||||
| LCNM | 5 | 10,790,853 | |||||
| SWLM | 5 | 10,958,834 | |||||
| Qcl-8 | 4 | 11,220,208 | 11,670,895 | LCNM | 4 | 11,649,295 | |
| WMM | 4 | 11,658,278 | |||||
| TSW | 4 | 11,661,092 | |||||
| Qcl-12 | 11 | 14,935,946 | 15,400,039 | LLM | 11 | 14,957,580 | |
| LLM | 11 | 15,003,280 | |||||
| TMM | 11 | 15,137,600 | |||||
| WMM | 11 | 15,137,600 | |||||
| CPMM | 11 | 15,138,140 | |||||
| LLM | 11 | 15,200,435 | |||||
| LMM | 11 | 15,219,964 | |||||
| LMM | 11 | 15,239,947 | |||||
| LMM | 11 | 15,289,738 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, R.; Dossa, K.; Li, D.; Yu, J.; You, J.; Wei, X.; Zhang, X. Genome-Wide Association Studies of 39 Seed Yield-Related Traits in Sesame (Sesamum indicum L.). Int. J. Mol. Sci. 2018, 19, 2794. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19092794
Zhou R, Dossa K, Li D, Yu J, You J, Wei X, Zhang X. Genome-Wide Association Studies of 39 Seed Yield-Related Traits in Sesame (Sesamum indicum L.). International Journal of Molecular Sciences. 2018; 19(9):2794. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19092794
Chicago/Turabian StyleZhou, Rong, Komivi Dossa, Donghua Li, Jingyin Yu, Jun You, Xin Wei, and Xiurong Zhang. 2018. "Genome-Wide Association Studies of 39 Seed Yield-Related Traits in Sesame (Sesamum indicum L.)" International Journal of Molecular Sciences 19, no. 9: 2794. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms19092794