Current Status of Proteomic Technologies for Discovering and Identifying Gingival Crevicular Fluid Biomarkers for Periodontal Disease
Abstract
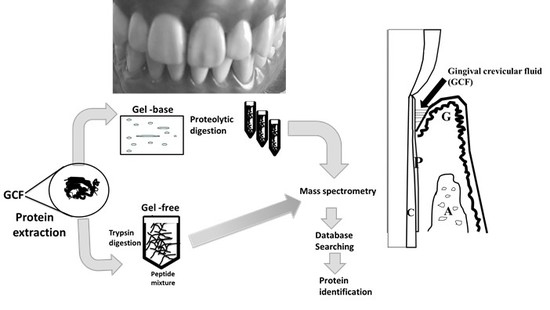
:1. Introduction
2. Human Gingival Crevicular Fluids (GCF) Samples
3. Discovery of Periodontal Disease Biomarkers by Proteomic Technology
3.1. Proteomic Technology (GC/MS, MALDI-TOF MS, and LC-MS/MS)
3.2. Labeling Methods in Mass Spectrometry Based on Quantitative Proteomics (SILAC, iTRAQ, and TMT)
4. Conclusions
Acknowledgments
Conflicts of Interest
Abbreviations
| MS | Mass spectrometry |
| MALDI-TOF MS | Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry |
| QTOF | Quadrupole TOF |
| ESI | Electrospray ionization |
| LC-MS/MS | Liquid chromatography (LC) coupled with tandem mass spectrometry (MS/MS) |
| SDS-PAGE | Sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| 2-DE | Two-dimensional electrophoresis |
References
- Genco, R.J.; Borgnakke, W.S. Risk factors for periodontal disease. Periodontol. 2000 2013, 62, 59–94. [Google Scholar] [CrossRef] [PubMed]
- Löe, H.; Silness, J. Periodontal disease in pregnancy. I. Prevalence and severity. Acta Odontol. Scand. 1963, 21, 533–551. [Google Scholar] [CrossRef] [PubMed]
- Löe, H. The Gingival Index, the plaque index and the Retention Index Systems. J. Periodontol. 1967, 38, 610–616. [Google Scholar] [CrossRef]
- Armitage, G.C. Development of a classification system for periodontal diseases and conditions. Ann. Periodontol. 1999, 4, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Izumi, Y.; Aoki, A.; Yamada, Y.; Kobayashi, H.; Iwata, T.; Akizuki, T.; Suda, T.; Nakamura, S.; Wara-Aswapati, N.; Ueda, M.; Ishikawa, I. Current and future periodontal tissue engineering. Periodontol. 2000 2011, 56, 166–187. [Google Scholar] [CrossRef] [PubMed]
- Listgarten, M.A. Pathogenesis of periodontitis. J. Clin. Periodontol. 1986, 13, 418–430. [Google Scholar] [CrossRef] [PubMed]
- Ebersole, J.L.; Dawson, D.R., 3rd; Morford, L.A.; Peyyala, R.; Miller, C.S.; Gonzaléz, O.A. Periodontal disease immunology: ‘Double indemnity’ in protecting the host. Periodontol. 2000 2013, 62, 163–202. [Google Scholar] [CrossRef]
- Chen, F.M.; Jin, Y. Periodontal tissue engineering and regeneration: Current approaches and expanding opportunities. Tissue Eng. Part B Rev. 2010, 16, 219–255. [Google Scholar] [CrossRef]
- Cochran, D.L.; Wozney, J.M. Biological mediators for periodontal regeneration. Periodontol. 2000 1999, 19, 40–58. [Google Scholar] [CrossRef]
- Listgarten, M.A. The role of dental plaque in gingivitis and periodontitis. J. Clin. Periodontol. 1988, 15, 485–487. [Google Scholar] [CrossRef]
- Schroeder, H.E.; Listgarten, M.A. The gingival tissues: The architecture of periodontal protection. Periodontol. 2000 1997, 13, 91–120. [Google Scholar] [CrossRef] [PubMed]
- Socransky, S.S.; Haffajee, A.D.; Cugini, M.A.; Smith, C.; Kent, R.L., Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998, 25, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Tribble, G.D.; Lamont, R.J. Bacterial invasion of epithelial cells and spreading in periodontal tissue. Periodontol. 2000 2010, 52, 68–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernández, M.; Dutzan, N.; García-Sesnich, J.; Abusleme, L.; Dezerega, A.; Silva, N.; González, F.E.; Vernal, R.; Sorsa, T.; Gamonal, J. Host-pathogen interactions in progressive chronic periodontitis. J. Dent. Res. 2011, 90, 1164–1170. [Google Scholar] [CrossRef] [PubMed]
- Megson, E.; Fitzsimmons, T.; Dharmapatni, K.; Bartold, P.M. C-reactive protein in gingival crevicular fluid may be indicative of systemic inflammation. J. Clin. Periodontol. 2010, 9, 797–804. [Google Scholar] [CrossRef] [PubMed]
- Kennett, C.N.; Cox, S.W.; Eley, B.M. Investigations into the cellular contribution to host tissue proteases and inhibitors in gingival crevicular fluid. J. Clin. Periodontol. 1997, 24, 424–431. [Google Scholar] [CrossRef] [PubMed]
- Adonogianaki, E.; Mooney, J.; Kinane, D.F. Detection of stable and active periodontitis sites by clinical assessment and gingival crevicular acute-phase protein levels. J. Periodontal Res. 1996, 31, 135–143. [Google Scholar] [CrossRef]
- Curtis, M.A.; Griffiths, G.S.; Price, S.J.; Coulthurst, S.K.; Johnson, N.W. The total protein concentration of gingival crevicular fluid. Variation with sampling time and gingival inflammation. J. Clin. Periodontol. 1988, 15, 628–632. [Google Scholar] [CrossRef]
- Bostanci, N.; Heywood, W.; Mills, K.; Parkar, M.; Nibali, L.; Donos, N. Application of label-free absolute quantitative proteomics in human gingival crevicular fluid by LC/MSE (gingival exudatome). J. Proteome. Res. 2010, 9, 2191–2199. [Google Scholar] [CrossRef]
- Bostanci, N.; Bao, K. Contribution of proteomics to our understanding of periodontal inflammation. Proteomics 2017, 17, 1500518. [Google Scholar] [CrossRef] [Green Version]
- Bostanci, N.; Belibasakis, G.N. Gingival crevicular fluid and its immune mediators in the proteomic era. Periodontol. 2000 2017, 76, 68–84. [Google Scholar] [CrossRef]
- Khurshid, Z.; Mali, M.; Naseem, M.; Najeeb, S.; Zafar, M.S. Human Gingival Crevicular Fluids (GCF) Proteomics: An Overview. Dent. J. 2017, 5, 12. [Google Scholar] [CrossRef] [PubMed]
- Cimasoni, G. Crevicular fluid updated. Monogr. Oral Sci. 1983, 12, 1–152. [Google Scholar]
- Challacombe, S.J.; Russell, M.W.; Hawkes, J. Passage of intact IgG from plasma to the oral cavity via crevicular fluid. Clin. Exp. Immunol. 1978, 34, 417–422. [Google Scholar]
- Lamster, I.B.; Ahlo, J.K. Analysis of gingival crevicular fluid as applied to the diagnosis of oral and systemic diseases. Ann. N. Y. Acad. Sci. 2007, 1098, 216–229. [Google Scholar] [CrossRef]
- Tözüm, T.F.; Hatipoğlu, H.; Yamalik, N.; Gürsel, M.; Alptekin, N.O.; Ataoğlu, T.; Marakoğlu, I.; Gürsoy, U.K.; Eratalay, K. Critical steps in electronic volume quantification of gingival crevicular fluid: The potential impact of evaporation, fluid retention, local conditions and repeated measurements. J. Periodontal Res. 2004, 39, 344–357. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, A. Methodological considerations in GCF sampling with paper strips: Poor recovery of uncomplexed elastase. J. Clin. Periodontol. 1996, 23, 432–436. [Google Scholar] [CrossRef]
- Preianò, M.; Falcone, D.; Maggisano, G.; Montalcini, T.; Navarra, M.; Paduano, S.; Savino, R.; Terracciano, R. Assessment of pre-analytical and analytical variables affecting peptidome profiling of gingival crevicular fluid by MALDI-TOF mass spectrometry. Clin. Chim. Acta 2014, 437, 120–128. [Google Scholar] [CrossRef]
- Preianò, M.; Maggisano, G.; Murfuni, M.S.; Villella, C.; Pelaia, C.; Montalcini, T.; Lombardo, N.; Pelaia, G.; Savino, R.; Terracciano, R. An Analytical Method for Assessing Optimal Storage Conditions of Gingival Crevicular Fluid and Disclosing a Peptide Biomarker Signature of Gingivitis by MALDI-TOF MS. Proteomics Clin. Appl. 2018, 12, 1800005. [Google Scholar] [CrossRef]
- Peakman, T.C.; Elliott, P. The UK Biobank sample handling and storage validation studies. Int. J. Epidemiol. 2008, 37 (Suppl. S1), i2–i6. [Google Scholar] [CrossRef]
- Elliott, P.; Peakman, T.C. The UK Biobank sample handling and storage protocol for the collection, processing and archiving of human blood and urine. Int. J. Epidemiol. 2008, 37, 234–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, D.H.; Kim, J.W.; Jeon, S.Y.; Park, B.K.; Han, B.G. Proteomic analysis of the effect of storage temperature on human serum. Ann. Clin. Lab. Sci. 2010, 40, 61–70. [Google Scholar] [PubMed]
- Nomura, F.; Tomonaga, T.; Sogawa, K.; Wu, D.; Ohashi, T. Application of proteomic technologies to discover and identify biomarkers for excessive alcohol consumption: A review. J. Chromatogr. B 2007, 855, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Nomura, F. Proteome-based bacterial identification using matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS): A revolutionary shift in clinical diagnostic microbiology. Biochim. Biophys. Acta 2015, 1854, 528–537. [Google Scholar] [CrossRef] [PubMed]
- Umemura, H.; Togawa, A.; Sogawa, K.; Satoh, M.; Mogushi, K.; Nishimura, M.; Matsushita, K.; Tanaka, H.; Takizawa, H.; Kodera, Y.; et al. Identification of a high molecular weight kininogen fragment as a marker for early gastric cancer by serum proteome analysis. J. Gastroenterol. 2011, 46, 577–585. [Google Scholar] [CrossRef] [PubMed]
- Satoh, M.; Haruta-Satoh, E.; Yamada, M.; Kado, S.; Nomura, F. Overexpression of hydroxymethylglutaryl CoA synthase 2 and 2,4-dienoyl-CoA reductase in rat pancreas following chronic alcohol consumption. Pancreas 2013, 42, 475–482. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Satoh, M.; Seimiya, M.; Sogawa, K.; Itoga, S.; Tomonaga, T.; Nomura, F. Combined proteomic analysis of liver tissue and serum in chronically alcohol-fed rats. Alcohol Clin. Exp. Res. 2013, 37, E79–E87. [Google Scholar] [CrossRef]
- Ogita, M.; Tsuchida, S.; Aoki, A.; Satoh, M.; Kado, S.; Sawabe, M.; Nanbara, H.; Kobayashi, H.; Takeuchi, Y.; Mizutani, K.; et al. Increased cell proliferation and differential protein expression induced by low-level Er:YAG laser irradiation in human gingival fibroblasts: Proteomic analysis. Lasers Med. Sci. 2015, 30, 1855–1866. [Google Scholar] [CrossRef]
- Tsuchida, S.; Satoh, M.; Umemura, H.; Sogawa, K.; Kawashima, Y.; Kado, S.; Sawai, S.; Nishimura, M.; Kodera, Y.; Matsushita, K.; Nomura, F. Proteomic analysis of gingival crevicular fluid for discovery of novel periodontal disease markers. Proteomics 2012, 12, 2190–2202. [Google Scholar] [CrossRef]
- Ngo, L.H.; Darby, I.B.; Veith, P.D.; Locke, A.G.; Reynolds, E.C. Mass spectrometric analysis of gingival crevicular fluid biomarkers can predict periodontal disease progression. J. Periodontal Res. 2012, 48, 331–341. [Google Scholar] [CrossRef]
- Sakellari, D. Proteomics of Periodontal Pocket. Curr. Oral Health Rep. 2017, 4, 271–277. [Google Scholar] [CrossRef]
- Batschkus, S.; Cingoez, G.; Urlaub, H.; Miosge, N.; Kirschneck, C.; Meyer-Marcotty, P.; Lenz, C. A new albumin-depletion strategy improves proteomic research of gingival crevicular fluid from periodontitis patients. Clin. Oral Investig. 2017, 22, 1375–1384. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Heo, S.H.; Lee, J.M.; Cho, J.Y. Identification of azurocidin as a potential periodontitis biomarker by a proteomic analysis of gingival crevicular fluid. Proteome Sci. 2011, 9, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eley, B.M.; Cox, S.W. Cathepsin B/L-, elastase-, tryptase-, trypsin- and dipeptidyl peptidase IV-like activities in gingival crevicular fluid: A comparison of levels before and after periodontal surgery in chronic periodontitis patients. J. Periodontol. 1992, 63, 412–417. [Google Scholar] [CrossRef] [PubMed]
- Travan, S.; Li, F.; D’Silva, N.J.; Slate, E.H.; Kirkwood, K.L. Differential expression of mitogen activating protein kinases in periodontitis. J. Clin. Periodontol. 2013, 40, 757–764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva-Boghossian, C.M.; Colombo, A.P.V.; Tanaka, M.; Rayo, C.; Xiao, Y.; Siqueira, W.L. Quantitative proteomic analysis of gingival crevicular fluid in different periodontal conditions. PLoS ONE 2013, 8, e75898. [Google Scholar] [CrossRef] [PubMed]
- Capelli, J.; Kantarci, A.; Haffajee, A.; Teles, R.P.; Fidel, R., Jr.; Figueredo, C.M. Matrix metalloproteinases and chemokines in the gingival crevicular fluid during orthodontic tooth movement. Eur. J. Orthod. 2011, 33, 705–711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozeki, M.; Nozaki, T.; Aoki, J.; Bamba, T.; Jensen, K.R.; Murakami, S.; Toyoda, M. Metabolomic Analysis of Gingival Crevicular Fluid Using Gas Chromatography/Mass Spectrometry. Mass Spectrom. 2016, 5, A0047. [Google Scholar] [CrossRef]
- Eijnde, B.O.; van Leemputte, M.; Brouns, F.; van Der Vuss, G.J.; Labarque, V.; Ramaekers, M.; Schuylenberg, R.; Verbessem, P.; Wijnen, L.T.; Hespel, P. No effects of oral ribose supplementation on repeated maximal exercise and de novo ATP resynthesis. J. Appl. Physiol. 2001, 91, 2275–2281. [Google Scholar] [CrossRef]
- Wei, Y.; Han, C.S.; Zhou, J.; Liu, Y.; Chen, L.; He, R.Q. D-ribose in glycation and protein aggregation. Biochim. Biophys. Acta 2012, 1820, 488–494. [Google Scholar] [CrossRef]
- Medzihradszky, K.F. Characterization of protein N-glycosylation. Methods Enzymol. 2005, 405, 116–138. [Google Scholar] [PubMed]
- Medzihradszky, K.F. Characterization of site-specific N-glycosylation. Methods Mol. Biol. 2008, 446, 293–316. [Google Scholar] [PubMed]
- Huynh, A.H.; Veith, P.D.; McGregor, N.R.; Adams, G.G.; Chen, D.; Reynolds, E.C.; Ngo, L.H.; Darby, I.B. Gingival crevicular fluid proteomes in health, gingivitis and chronic periodontitis. J. Periodontal Res. 2014, 50, 637–649. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Pradeep, A.R.; Raghavendra, N.M.; Arjun, P.; Kathariya, R. Gingival crevicular fluid and serum cystatin c levels in periodontal health and diseas. Dis. Markers 2012, 32, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Yaprak, E.; Kasap, M.; Akpınar, G.; Kayaaltı-Yüksek, S.; Sinanoğlu, A.; Guzel, N.; Kocasarac, H.D. The prominent proteins expressed in healthy gingiva: A pilot exploratory tissue proteomics study. Odontology 2018, 106, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Rody, W.J., Jr.; Holliday, L.S.; McHugh, K.P.; Wallet, S.M.; Spicer, V.; Krokhin, O. Mass spectrometry analysis of gingival crevicular fluid in the presence of external root resorption. Am. J. Orthod. Dentofac. Orthop. 2014, 145, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Tamura, T.; Hamachi, I. Chemical proteomics for subcellular proteome analysis. Curr. Opin. Chem. Biol. 2018, 48, 1–7. [Google Scholar] [CrossRef]
- Wright, M.H. Chemical Proteomics of Host-Microbe Interactions. Proteomics 2018, 18, e1700333. [Google Scholar] [CrossRef]
- Yang, Y.; Fonović, M.; Verhelst, S.H. Cleavable Linkers in Chemical Proteomics Applications. Methods Mol. Biol. 2017, 1491, 185–203. [Google Scholar]
- Wang, J.; Gao, L.; Lee, Y.M.; Kalesh, K.A.; Ong, Y.S.; Lim, J.; Jee, J.E.; Sun, H.; Lee, S.S.; Hua, Z.C.; et al. Target identification of natural and traditional medicines with quantitative chemical proteomicsapproaches. Pharmacol. Ther. 2016, 162, 10–22. [Google Scholar] [CrossRef]
- Chahrour, O.; Cobice, D.; Malone, J. Stable isotope labelling methods in mass spectrometry-based quantitative proteomics. J. Pharm. Biomed. Anal. 2015, 113, 2–20. [Google Scholar] [CrossRef] [PubMed]
- Wright, M.H.; Sieber, S.A. Chemical proteomics approaches for identifying the cellular targets of natural products. Nat. Prod. Rep. 2016, 33, 681–708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terzi, F.; Cambridge, S. An Overview of Advanced SILAC-Labeling Strategies for Quantitative Proteomics. Methods Enzymol. 2017, 585, 29–47. [Google Scholar] [PubMed]
- Dittmar, G.; Selbach, M. SILAC for biomarker discovery. Proteomics Clin. Appl. 2015, 9, 301–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mann, M. Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell Biol. 2006, 7, 952–958. [Google Scholar] [CrossRef] [PubMed]
- Wiese, S.; Reidegeld, K.A.; Meyer, H.E.; Warscheid, B. Protein labeling by iTRAQ: A new tool for quantitative mass spectrometry in proteome research. Proteomics 2007, 7, 340–350. [Google Scholar] [CrossRef] [PubMed]
- Dayon, L.; Turck, N.; Garcí-Berrocoso, T.; Walter, N.; Burkhard, P.R.; Vilalta, A.; Sahuquillo, J.; Montaner, J.; Sanchez, J.C. Brain extracellular fluid protein changes in acute stroke patients. J. Proteome Res. 2011, 10, 1043–1051. [Google Scholar] [CrossRef] [PubMed]
- Arntzen, M.Ø.; Koehler, C.J.; Barsnes, H.; Berven, F.S.; Treumann, A.; Thiede, B. IsobariQ: Software for isobaric quantitative proteomics using IPTL, iTRAQ, and TMT. J. Proteome Res. 2011, 10, 913–920. [Google Scholar] [CrossRef]
- Giron, P.; Dayon, L.; Turck, N.; Hoogland, C.; Sanchez, J.C. Quantitative analysis of human cerebrospinal fluid proteins using a combination of cysteine tagging and amine-reactive isobaric labeling. J. Proteome Res. 2011, 10, 249–258. [Google Scholar] [CrossRef]
- Baeumlisberger, D.; Arrey, T.N.; Rietschel, B.; Rohmer, M.; Papasotiriou, D.G.; Mueller, B.; Beckhaus, T.; Karas, M. Labeling elastase digests with TMT: Informational gain by identification of poorly detectable peptides with MALDI-TOF/TOF mass spectrometry. Proteomics 2010, 10, 3905–3909. [Google Scholar] [CrossRef]
- Dayon, L.; Turck, N.; Kienle, S.; Schulz-Knappe, P.; Hochstrasser, D.F.; Scherl, A.; Sanchez, J.C. Isobaric tagging-based selection and quantitation of cerebrospinal fluid tryptic peptides with reporter calibration curves. Anal. Chem. 2010, 82, 848–858. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, S.; Satoh, M.; Kawashima, Y.; Sogawa, K.; Kado, S.; Sawai, S.; Nishimura, M.; Ogita, M.; Takeuchi, Y.; Kobyashi, H.; et al. Application of quantitative proteomic analysis using tandem mass tags for discovery and identification of novel biomarkers in periodontal disease. Proteomics 2013, 13, 2339–2350. [Google Scholar] [CrossRef] [PubMed]
- Moriya, Y.; Obama, T.; Aiuchi, T.; Sugiyama, T.; Endo, Y.; Koide, Y.; Noguchi, E.; Ishizuka, M.; Inoue, M.; Itabe, H.; et al. Quantitative proteomic analysis of gingival crevicular fluids from deciduous and permanent teeth. J. Clin. Periodontol. 2017, 44, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, L.G.; Nouh, H.; Salih, E. Quantitative gingival crevicular fluid proteome in health and periodontal disease using stable isotope chemistries and mass spectrometry. J. Clin. Periodontol. 2014, 41, 733–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Study | Methodology | Number of Identified Proteins | Candidate Protein Biomarkers of Periodontal Disease | Ref. |
|---|---|---|---|---|
| Carina et al., 2013 | Gel-free: LC-MS/MS | 230 proteins | Immune responses as Ig gamma-1 chain C region, Ig gamma-3 chain C region, Lactoferroxin-C, Lactrotransferrin, Leukocyte elastase inhibitor, Apolipoprotein E, Alpha-1 antitrypsin, Annexin, Cathelicidin antimicrobial peptide, Cathepsin G, Coronin-1A, Dermcidin isoform 2, Heat shock protein beta-1, Myeloperoxidase, Neutrophil defensin 3, S100 A8, S100 A9, Myosin 9, Annexin A1 | [46] |
| Wellington et al., 2014 | Gel-free: LC-MS/MS | 2789 proteins | Exosomal proteins | [56] |
| Leandro et al., 2014 | Gel-free: LC-MS/MS, stable isotope-labeling reagents, isotope-coded affinity tag, mTRAQ | 238 proteins | Host-derived Ig α-2 chain C, Kallikrein-4, S100A9, Transmembrane proteinase 13, Peptidase S1 domain, Collagens, Pathogenic bacterial proteins | [74] |
| Huynh et al., 2014 | 1D-PAGE: LC-MS/MS | 121 proteins | Cystatins B and S, Immunoglobulins, Keratin proteins, Fibronectin, Lactotransferrin precursor, 14-3-3 protein zeta/delta, Neutrophil defensin 3, α-actinin | [53] |
| Ozeki et al., 2016 | Gel-free: GC/MS | 19 metabolites | Putrescine, Lysine, Phenylalanine, Ribose, Taurine, 5-aminovaleric acid, Galactose | [48] |
| Moriya et al., 2017 | Gel-free: LC-MS/MS, iTRAQ technique | 62 proteins | Neutrophil-derived proteins, Myeloperoxidase, Lactoferrin, | [73] |
| Batschkus et al., 2018 | 1D-PAGE: LC-MS/MS | 317 proteins | Azurocidin, Cathepsin B, Mitogen-activated protein kinases, Coronin 1A, MMP-9 | [42] |
| Yaprak et al., 2018 | 2-DE; MALDI-TOF/TOF | 47 proteins | 14-3-3 protein sigma, Protein DJ-1, α-enolase, Triosephosphate isomerase, Superoxide dismutase, Peroxiredoxin-1, Protein S100-A9, Galectin-7, Annexin A2/A4, Carbonic anhydrase 1, Chaperone proteins | [55] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tsuchida, S.; Satoh, M.; Takiwaki, M.; Nomura, F. Current Status of Proteomic Technologies for Discovering and Identifying Gingival Crevicular Fluid Biomarkers for Periodontal Disease. Int. J. Mol. Sci. 2019, 20, 86. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20010086
Tsuchida S, Satoh M, Takiwaki M, Nomura F. Current Status of Proteomic Technologies for Discovering and Identifying Gingival Crevicular Fluid Biomarkers for Periodontal Disease. International Journal of Molecular Sciences. 2019; 20(1):86. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20010086
Chicago/Turabian StyleTsuchida, Sachio, Mamoru Satoh, Masaki Takiwaki, and Fumio Nomura. 2019. "Current Status of Proteomic Technologies for Discovering and Identifying Gingival Crevicular Fluid Biomarkers for Periodontal Disease" International Journal of Molecular Sciences 20, no. 1: 86. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20010086