Effects of KEAP1 Silencing on the Regulation of NRF2 Activity in Neuroendocrine Lung Tumors
Abstract
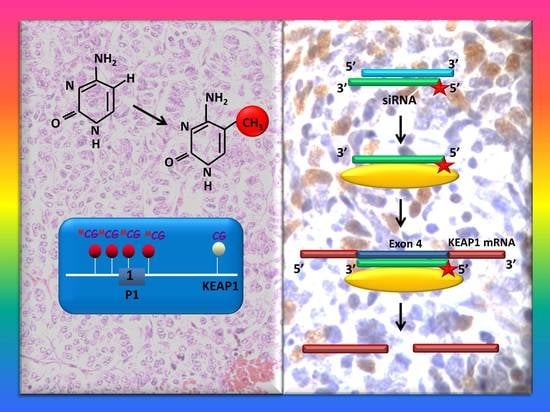
:1. Background
2. Results
2.1. The KEAP1 Silencing Affects the NRF2 and Expression Levels of TXNRD1, AKR1C1 and NQO1 in Carcinoid Lines
2.2. Restoration of KEAP1 Expression Correlates with KEAP1 P1 Region Demethylation by 5-aza-dC Treatment in Carcinoid Cell Lines
2.3. KEAP1 Promoter Region Hypermethylation are Frequent Epigenetic in Lung Carcinoids, Whereas Point Mutations are Absent
3. Discussion
4. Methods
4.1. Cell Lines
4.2. KEAP1 Silencing by siRNA
4.3. Western Blot Analysis
4.4. In Vitro 5-Aza-2′-deoxycytidine (5-aza-dC) Treatment
4.5. RNA Extraction, Reverse Transcription and Quantitative Real-Time PCR (qRT-PCR)
4.6. Patients and Tissue Samples
4.7. Bisulfite Conversion and Quantitative Methylation Specific-PCR Analysis (qMSP)
4.8. Mutation Analysis And Loss of Heterozygosity Analysis (LOH)
4.9. Immunohistochemistry (IHC)
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kitamura, H.; Motohashi, H. NRF2 addiction in cancer cells. Cancer Sci. 2018, 109, 900–911. [Google Scholar] [CrossRef] [Green Version]
- Fabrizio, F.P.; Sparaneo, A.; Trombetta, D.; Muscarella, L.A. Epigenetic versus Genetic Deregulation of the KEAP1/NRF2 Axis in Solid Tumors: Focus on Methylation and Noncoding RNAs. Oxid. Med. Cell. Longev. 2018, 2018, 2492063. [Google Scholar] [CrossRef]
- Muscarella, L.A.; Parrella, P.; D’Alessandro, V.; la Torre, A.; Barbano, R.; Fontana, A.; Tancredi, A.; Guarnieri, V.; Balsamo, T.; Coco, M.; et al. Frequent epigenetics inactivation of KEAP1 gene in non-small cell lung cancer. Epigenetics 2011, 6, 710–719. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.; An, J.; Ji, F.; Jiao, H.; Sun, H.; Zhou, D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem. Biophys. Res. Commun. 2008, 373, 151–154. [Google Scholar] [CrossRef]
- Frank, R.; Scheffler, M.; Merkelbach-Bruse, S.; Ihle, M.A.; Kron, A.; Rauer, M.; Ueckeroth, F.; König, K.; Michels, S.; Fischer, R.; et al. Clinical and Pathological Characteristics of KEAP1- and NFE2L2-Mutated Non-Small Cell Lung Carcinoma (NSCLC). Clin. Cancer Res. 2018, 24, 3087–3096. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Misra, V.; Thimmulappa, R.K.; Lee, H.; Ames, S.; Hoque, M.O.; Herman, J.G.; Baylin, S.B.; Sidransky, D.; Gabrielson, E.; et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006, 3, e420. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Cuesta, L.; Peifer, M.; Lu, X.; Seidel, D.; Zander, T.; Leenders, F.; Luka, O.; Brustugun, O.-T.; Field, J.K.; Wright, G.; et al. 1531: Crossentity mutation analysis of lung neuroendocrine tumors sheds light into their molecular origin and identifies new therapeutic targets. Cancer Res. 2014. [Google Scholar] [CrossRef]
- Fernandez-Cuesta, L.; Peifer, M.; Lu, X.; Sun, R.; Ozretić, L.; Seidal, D.; Zander, T.; Leenders, F.; George, J.; Müller, C.; et al. Frequent mutations in chromatin-remodeling genes in pulmonary carcinoids. Nat. Commun. 2014, 5, 3518. [Google Scholar] [CrossRef]
- Simbolo, M.; Mafficini, A.; Sikora, K.O.; Fassan, M.; Barbi, S.; Corbo, V.; Mastracci, L.; Rusev, B.; Grillo, F.; Vicentini, C.; et al. Lung neuroendocrine tumors: Deep sequencing of the four World Health Organization histotypes reveals chromatin-remodeling genes as major players and a prognostic role for TERT, RB1, MEN1 and KMT2D. J. Pathol. 2017, 241, 488–500. [Google Scholar] [CrossRef]
- Karpathakis, A.; Dibra, H.; Thirlwell, C. Neuroendocrine tumors: Cracking the epigenetic code. Endocr. Relat. Cancer 2013, 20, R65–R82. [Google Scholar] [CrossRef] [PubMed]
- Patel, P.; Galoian, K. Molecular challenges of neuroendocrine tumors. Oncol. Lett. 2018, 15, 2715–2725. [Google Scholar] [CrossRef] [PubMed]
- Derks, J.L.; Leblay, N.; Lantuejoul, S.; Dingemans, A.C.; Speel, E.M.; Fernandez-Cuesta, L. New Insights into the Molecular Characteristics of Pulmonary Carcinoids and Large Cell Neuroendocrine Carcinomas, and the Impact on Their Clinical Management. J. Thorac. Oncol. 2018, 13, 752–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rossi, G.; Bertero, L.; Marchiò, C.; Papotti, M. Molecular alterations of neuroendocrine tumors of the lung. Histopathology 2018, 72, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, G.; Fumagalli, C.; Trubia, M.; Sonzogni, A.; Rekhtman, N.; Maisonneuve, P.; Galetta, D.; Spaggiari, L.; Veronesi, G.; Scarpa, A.; et al. Dual role of RASSF1 as a tumor suppressor and an oncogene in neuroendocrine tumors of the lung. Anticancer Res. 2010, 30, 4269–4281. [Google Scholar] [PubMed]
- La Torre, A.; Muscarella, L.A.; Parrella, P.; Balsamo, T.; Bisceglia, M.; Valori, V.M.; la Torre, A.; Barbano, R.; Perrella, E.; Poeta, M.L.; et al. Aberrant genes promoter methylation in neural crest-derived tumors. Int. J. Biol. Markers 2012, 27, e389–e394. [Google Scholar] [CrossRef] [PubMed]
- Chaussade, L.; Eymin, B.; Brambilla, E.; Gazzeri, S. Expression of p15 and p15.5 products in neuroendocrine lung tumors: Relationship with p15(INK4b) methylation status. Oncogene 2001, 20, 6587–6596. [Google Scholar] [CrossRef] [PubMed]
- Menegon, S.; Columbano, A.; Giordano, S. The Dual Roles of NRF2 in Cancer. Trends Mol. Med. 2016, 22, 578–593. [Google Scholar] [CrossRef]
- Hayes, J.D.; Dinkova-Kostova, A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014, 39, 199–218. [Google Scholar] [CrossRef]
- Levings, D.C.; Wang, X.; Kohlhase, D.; Bell, D.A.; Slattery, M. A distinct class of antioxidant response elements is consistently activated in tumors with NRF2 mutations. Redox Biol. 2018, 19, 235–249. [Google Scholar] [CrossRef]
- Tanca, A.; Addis, M.F.; Pagnozzi, D.; Cossu-Rocca, P.; Tonelli, R.; Falchi, G.; Eccher, A.; Roggio, T.; Fanciulli, G.; Uzzau, S. Proteomic analysis of formalin-fixed, paraffin-embedded lung neuroendocrine tumor samples from hospital archives. J. Proteom. 2011, 74, 359–370. [Google Scholar] [CrossRef]
- Bloom, D.; Dhakshinamoorthy, S.; Jaiswal, A.K. Site-directed mutagenesis of cysteine to serine in the DNA binding region of Nrf2 decreases its capacity to upregulate antioxidant response element-mediated expression and antioxidant induction of NAD(P)H:quinone oxidoreductase1 gene. Oncogene 2002, 21, 2191–2200. [Google Scholar] [CrossRef]
- Wang, X.J.; Hayes, J.D.; Wolf, C.R. Generation of a stable antioxidant response element-driven reporter gene cell line and its use to show redox-dependent activation of nrf2 by cancer chemotherapeutic agents. Cancer Re. 2006, 66, 10983–10994. [Google Scholar] [CrossRef]
- Kerins, M.J.; Ooi, A. The Roles of NRF2 in Modulating Cellular Iron Homeostasis. Antioxid Redox Signal 2017, 29, 1756–1773. [Google Scholar] [CrossRef]
- Tanca, A.; Addis, M.F.; Pisanu, S.; Abbondio, M.; Pagnozzi, D.; Eccher, A.; Rindi, G.; Cossu-Rocca, P.; Uzzau, S.; Fanciulli, G. Atypical carcinoid and large cell neuroendocrine carcinoma of the lung: A proteomic dataset from formalin-fixed archival samples. Data Brief 2016, 7, 529–531. [Google Scholar] [CrossRef]
- Sakai, C.; Ishida, M.; Ohba, H.; Yamashita, H.; Uchida, H.; Yoshizumi, M.; Ishida, T. Fish oil omega-3 polyunsaturated fatty acids attenuate oxidative stress-induced DNA damage in vascular endothelial cells. PLoS ONE 2017, 12, e0187934. [Google Scholar] [CrossRef]
- Kato, K.; Takahashi, K.; Monzen, S.; Yamamoto, H.; Maruyama, A.; Itoh, K.; Kashiwakura, I. Relationship between radiosensitivity and Nrf2 target gene expression in human hematopoietic stem cells. Radiat. Res. 2010, 174, 177–184. [Google Scholar] [CrossRef]
- Pietsch, E.C.; Chan, J.Y.; Torti, F.M.; Torti, S.V. Nrf2 mediates the induction of ferritin H in response to xenobiotics and cancer chemopreventive dithiolethiones. J. Biol. Chem. 2003, 278, 2361–2369. [Google Scholar] [CrossRef]
- Qaisiya, M.; Coda Zabetta, C.D.; Bellarosa, C.; Tiribelli, C. Bilirubin mediated oxidative stress involves antioxidant response activation via Nrf2 pathway. Cell Signal 2014, 26, 512–520. [Google Scholar] [CrossRef]
- Sun, X.; Ou, Z.; Chen, R.; Niu, X.; Chen, D.; Kang, R.; Tang, D. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology 2016, 63, 173–184. [Google Scholar] [CrossRef]
- Li, N.; Alam, J.; Venkatesan, M.I.; Eiguren-Fernandez, A.; Schmitz, D.; Di Stefano, E.; Slaughter, N.; Killeen, E.; Wang, X.; Huang, A.; et al. Nrf2 is a key transcription factor that regulates antioxidant defense in macrophages and epithelial cells: Protecting against the proinflammatory and oxidizing effects of diesel exhaust chemicals. J. Immunol. 2004, 173, 3467–3481. [Google Scholar] [CrossRef]
- Molatlhegi, R.P.; Phulukdaree, A.; Anand, K.; Gengan, R.M.; Tiloke, C.; Chuturgoon, A.A. Cytotoxic Effect of a Novel Synthesized Carbazole Compound on A549 Lung Cancer Cell Line. PLoS ONE 2015, 10, e0129874. [Google Scholar] [CrossRef]
- Son, Y.O.; Pratheeshkumar, P.; Roy, R.V.; Hitron, J.A.; Wang, L.; Zhang, Z.; Shi, X. Nrf2/p62 signaling in apoptosis resistance and its role in cadmium-induced carcinogenesis. J. Biol. Chem. 2014, 289, 28660–28675. [Google Scholar] [CrossRef]
- Yao, W.; Luo, G.; Zhu, G.; Chi, X.; Zhang, A.; Xia, Z.; Hei, Z. Propofol activation of the Nrf2 pathway is associated with amelioration of acute lung injury in a rat liver transplantation model. Oxid. Med. Cell. Longev. 2014, 258567. [Google Scholar] [CrossRef]
- Zhang, R.; Chae, S.; Lee, J.H.; Hyun, J.W. The cytoprotective effect of butin against oxidative stress is mediated by the up-regulation of manganese superoxide dismutase expression through a PI3K/Akt/Nrf2-dependent pathway. J. Cell Biochem. 2012, 113, 1987–1997. [Google Scholar] [CrossRef]
- Zhu, H.; Itoh, K.; Yamamoto, M.; Zweier, J.L.; Li, Y. Role of Nrf2 signaling in regulation of antioxidants and phase 2 enzymes in cardiac fibroblasts: Protection against reactive oxygen and nitrogen species-induced cell injury. FEBS Lett. 2005, 579, 3029–3036. [Google Scholar] [CrossRef] [Green Version]
- Hyeon, S.; Lee, H.; Yang, Y.; Jeong, W. Nrf2 deficiency induces oxidative stress and promotes RANKL-induced osteoclast differentiation. Free Radic. Biol. Med. 2013, 65, 789–799. [Google Scholar] [CrossRef]
- Liu, R.; Chen, H.; Bai, H.; Zhang, W.; Wang, X.; Qin, X.; Zhang, X.; Li, W.; Liang, X.; Hai, C. Suppression of nuclear factor erythroid 2-related factor 2 via extracellular signal-regulated kinase contributes to bleomycin-induced oxidative stress and fibrogenesis. Toxicol. Lett. 2013, 220, 15–25. [Google Scholar] [CrossRef]
- Noma, N.; Fujii, G.; Miyamoto, S.; Komiya, M.; Nakanishi, R.; Shimura, M.; Tanuma, S.I.; Mutoh, M. Impact of Acetazolamide, a Carbonic Anhydrase Inhibitor, on the Development of Intestinal Polyps in Min Mice. Int. J. Mol. Sci. 2017, 18, 851. [Google Scholar] [CrossRef]
- Ashino, T.; Yamamoto, M.; Numazawa, S. Nrf2/Keap1 system regulates vascular smooth muscle cell apoptosis for vascular homeostasis: Role in neointimal formation after vascular injury. Sci. Rep. 2016, 6, 26291. [Google Scholar] [CrossRef]
- Lee, Y.J.; Lee, D.M.; Lee, S.H. Nrf2 Expression and Apoptosis in Quercetin-treated Malignant Mesothelioma Cells. Mol. Cells 2015, 38, 416–425. [Google Scholar] [CrossRef] [Green Version]
- Cheng, M.L.; Lu, Y.F.; Chen, H.; Shen, Z.Y.; Liu, J. Liver expression of Nrf2-related genes in different liver diseases. Hepatobiliary Pancreat. Dis. Int. 2015, 14, 485–491. [Google Scholar] [CrossRef]
- Matte, A.; De Falco, L.; Iolascon, A.; Mohandas, N.; An, X.; Siciliano, A.; Leboeuf, C.; Janin, A.; Bruno, M.; Choi, S.Y.; et al. The Interplay Between Peroxiredoxin-2 and Nuclear Factor-Erythroid 2 Is Important in Limiting Oxidative Mediated Dysfunction in beta-Thalassemic Erythropoiesis. Antioxid Redox Signal 2015, 23, 1284–1297. [Google Scholar] [CrossRef] [PubMed]
- Olahova, M.; Veal, E.A. A peroxiredoxin, PRDX-2, is required for insulin secretion and insulin/IIS-dependent regulation of stress resistance and longevity. Aging Cell 2015, 14, 558–568. [Google Scholar] [CrossRef]
- Wang, T.; Liang, X.; Abeysekera, I.R.; Iqbal, U.; Duan, Q.; Naha, G.; Lin, L.; Yao, X. Activation of the Nrf2-Keap 1 Pathway in Short-Term Iodide Excess in Thyroid in Rats. Oxid. Med. Cell. Longev. 2017, 2017, 4383652. [Google Scholar] [CrossRef]
- Kim, Y.S.; Lee, H.L.; Lee, K.B.; Park, J.H.; Chung, W.Y.; Lee, K.S.; Sheen, S.S.; Park, K.J.; Hwang, S.C. Nuclear factor E2-related factor 2 dependent overexpression of sulfiredoxin and peroxiredoxin III in human lung cancer. Korean J. Intern. Med. 2011, 26, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Liverman, C.S.; Cui, L.; Yong, C.; Choudhuri, R.; Klein, R.M.; Welch, K.M.; Berman, N.E. Response of the brain to oligemia: Gene expression, c-Fos, and Nrf2 localization. Brain Res. Mol. Brain Res. 2004, 126, 57–66. [Google Scholar] [CrossRef]
- Belcher, J.D.; Chen, C.; Nguyen, J.; Zhang, P.; Abdulla, F.; Nguyen, P.; Killeen, T.; Xu, P.; O’Sullivan, G.; Nath, K.A.; et al. Control of Oxidative Stress and Inflammation in Sickle Cell Disease with the Nrf2 Activator Dimethyl Fumarate. Antioxid Redox Signal 2017, 26, 748–762. [Google Scholar] [CrossRef]
- Gallorini, M.; Petzel, C.; Bolay, C.; Hiller, K.A.; Cataldi, A.; Buchalla, W.; Krifka, S.; Schweikl, H. Activation of the Nrf2-regulated antioxidant cell response inhibits HEMA-induced oxidative stress and supports cell viability. Biomaterials 2015, 56, 114–128. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Abdo, S.; Zhao, S.; Wu, C.H.; Shi, Y.; Lo, C.S.; Chenier, I.; Alquier, T.; Filep, J.G.; Ingelfinger, J.R.; et al. Insulin Inhibits Nrf2 Gene Expression via Heterogeneous Nuclear Ribonucleoprotein F/K in Diabetic Mice. Endocrinology 2017, 158, 903–919. [Google Scholar] [CrossRef] [Green Version]
- Moujalled, D.; Grubman, A.; Acevedo, K.; Yang, S.; Ke, Y.D.; Moujalled, D.M.; Duncan, C.; Caragounis, A.; Perera, N.D.; Turner, B.J.; et al. TDP-43 mutations causing amyotrophic lateral sclerosis are associated with altered expression of RNA-binding protein hnRNP K and affect the Nrf2 antioxidant pathway. Hum. Mol. Genet. 2017, 26, 1732–1746. [Google Scholar] [CrossRef]
- Huang, B.W.; Ray, P.D.; Iwasaki, K.; Tsuji, Y. Transcriptional regulation of the human ferritin gene by coordinated regulation of Nrf2 and protein arginine methyltransferases PRMT1 and PRMT4. FASEB J. 2013, 27, 3763–3774. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Li, H.; Liu, L.; Lu, Y.; Gao, Y.; Geng, P.; Li, X.; Huang, B.; Zhang, Y.; Lu, J. Methylation of arginine by PRMT1 regulates Nrf2 transcriptional activity during the antioxidative response. Biochim. Biophys. Acta. 2016, 1863, 2093–2103. [Google Scholar] [CrossRef]
- Guo, D.; Wu, B.; Yan, J.; Li, X.; Sun, H.; Zhou, D. A possible gene silencing mechanism: Hypermethylation of the Keap1 promoter abrogates binding of the transcription factor Sp1 in lung cancer cells. Biochem. Biophys. Res. Commun. 2012, 428, 80–85. [Google Scholar] [CrossRef]
- Wang, Q.; Li, J.; Yang, X.; Sun, H.; Gao, S.; Zhu, H.; Wu, J.; Jin, W. Nrf2 is associated with the regulation of basal transcription activity of the BRCA1 gene. Acta Biochim. Biophys. Sin. (Shanghai) 2013, 45, 179–187. [Google Scholar] [CrossRef] [Green Version]
- Lazaro, I.; Oguiza, A.; Recio, C.; Lopez-Sanz, L.; Bernal, S.; Egido, J.; Gomez-Guerrero, C. Interplay between HSP90 and Nrf2 pathways in diabetes-associated atherosclerosis. Clin. Investig Arter. 2017, 29, 51–59. [Google Scholar]
- Niture, S.K.; Jaiswal, A.K. Hsp90 interaction with INrf2(Keap1) mediates stress-induced Nrf2 activation. J. Biol. Chem. 2010, 285, 36865–36875. [Google Scholar] [CrossRef]
- Sahin, K.; Orhan, C.; Akdemir, F.; Tuzcu, M.; Iben, C.; Sahin, N. Resveratrol protects quail hepatocytes against heat stress: Modulation of the Nrf2 transcription factor and heat shock proteins. J. Anim. Physiol. Anim. Nutr. (Berl) 2012, 96, 66–74. [Google Scholar] [CrossRef]
- Lee, S.C.; Zhang, J.; Strom, J.; Yang, D.; Dinh, T.N.; Kappeler, K.; Chen, Q.M. G-Quadruplex in the NRF2 mRNA 5′ Untranslated Region Regulates De Novo NRF2 Protein Translation under Oxidative Stress. Mol. Cell Biol. 2016, 37, e00122-16. [Google Scholar] [CrossRef]
- Hendifar, A.E.; Marchevsky, A.M.; Tuli, R. Neuroendocrine Tumors of the Lung: Current Challenges and Advances in the Diagnosis and Management of Well-Differentiated Disease. J. Thorac. Oncol. 2017, 12, 425–436. [Google Scholar] [CrossRef] [Green Version]
- Derks, J.; Leblay, N.; van Suylen, R.J.; Thunnissen, E.; den Bakker, M.; Groen, H.J.M.; Smit, E.F.; Damhuis, R.; van de Broek, E.; Chabrier, A.; et al. Genetic subtypes of large cell neuroendocrine carcinoma (LCNEC) to predict response to chemotherapy. J. Clin. Oncol. 2017, 35, 9061. [Google Scholar] [CrossRef]
- Mitsuishi, Y.; Motohashi, H.; Yamamoto, M. The Keap1-Nrf2 system in cancers: Stress response and anabolic metabolism. Front. Oncol. 2012, 2, 200. [Google Scholar] [CrossRef]
- Wondrak, G.T. Redox-directed cancer therapeutics: Molecular mechanisms and opportunities. Antioxid Redox Signal 2009, 11, 3013–3069. [Google Scholar] [CrossRef]
- Bendavit, G.; Aboulkassim, T.; Hilmi, K.; Shah, S.; Batist, G. Nrf2 Transcription Factor Can Directly Regulate mTOR:linking cytoprotective gene expression to a major metaboli regulator that generates redox activity. J. Biol. Chem. 2016, 291, 25476–25488. [Google Scholar] [CrossRef]
- Sancak, Y.; Peterson, T.R.; Shaul, Y.D.; Lindquist, R.A.; Thoreen, C.C.; Bar-Peled, L.; Sabatini, D.M. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 2008, 320, 1496–1501. [Google Scholar] [CrossRef]
- Chan, J.; Kulke, M. Targeting the mTOR signaling pathway in neuroendocrine tumors. Curr. Treat. Options Oncol. 2014, 15, 365–379. [Google Scholar] [CrossRef]
- Cescon, D.W.; She, D.; Sakashita, S.; Zhu, C.Q.; Pintilie, M.; Shepherd, F.A.; Tsao, M.S. NRF2 Pathway Activation and Adjuvant Chemotherapy Benefit in Lung Squamous Cell Carcinoma. Clin. Cancer Res. 2015, 21, 2499–2505. [Google Scholar] [CrossRef] [Green Version]
- Bryan, H.K.; Olayanju, A.; Goldring, C.E.; Park, B.K. The Nrf2 cell defence pathway: Keap1-dependent and -independent mechanisms of regulation. Biochem. Pharmacol. 2013, 85, 705–717. [Google Scholar] [CrossRef]
- Ren, D.; Villeneuve, N.F.; Jiang, T.; Wu, T.; Lau, A.; Toppin, H.A.; Zhang, D.D. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc. Natl. Acad. Sci. USA 2011, 108, 1433–1438. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.; Wang, H.; Fan, L.; Wu, X.; Xin, A.; Ren, H.; Wang, X.J. Luteolin inhibits Nrf2 leading to negative regulation of the Nrf2/ARE pathway and sensitization of human lung carcinoma A549 cells to therapeutic drugs. Free Radic. Biol. Med. 2011, 50, 1599–1609. [Google Scholar] [CrossRef]
- Magesh, S.; Chen, Y.; Hu, L. Small molecule modulators of Keap1– Nrf2–ARE pathway as potential preventive and therapeutic agents. Med. Res. Rev. 2012, 32, 687–726. [Google Scholar] [CrossRef]
- Singh, A.; Venkannagari, S.; Oh, K.H.; Zhang, Y.Q.; Rohde, J.M.; Liu, L.; Nimmagadda, S.; Sudini, K.; Brimacombe, K.R.; Gajghate, S.; et al. Small molecule inhibitor of NRF2 selectively intervenes therapeutic resistance in KEAP1-deficient NSCLC tumors. ACS Chem. Biol. 2016, 11, 3214–3225. [Google Scholar] [CrossRef]
- Choi, E.J.; Jung, B.J.; Lee, S.H.; Yoo, H.S.; Shin, E.A.; Ko, H.J.; Chang, S.; Kim, S.Y.; Jeon, S.M. A clinical drug library screen identifies clobetasol propionate as an NRF2 inhibitor with potential therapeutic efficacy in KEAP1 mutant lung cancer. Oncogene 2017, 36, 5285–5295. [Google Scholar] [CrossRef]
- Muscarella, L.A.; Barbano, R.; D’Angelo, V.; Copetti, M.; Coco, M.; Balsamo, T.; la Torre, A.; Notarangelo, A.; Troiano, M.; Parisi, S.; et al. Regulation of KEAP1 expression by promoter methylation in malignant gliomas and association with patient’s outcome. Epigenetics 2011, 6, 317–325. [Google Scholar] [CrossRef]





| ID Sample | Histotypes | Gender | KEAP1 (Meth and/or LOH 1) | Years at Diagnosis | UICC Stage | Progression (Y/N) | Status (D/A) | DFS (mos) | OS (mos) |
|---|---|---|---|---|---|---|---|---|---|
| LCCH01-322 | AC | M | Meth | 52 | IIA | Y | A | 84 | 120 |
| LCCH01-328 | AC | M | LOH | 47 | IIA | N | A | 67 | 67 |
| LCCH01-329 | AC | M | LOH | 49 | IA | N | A | 124 | 204 |
| LCCH01-330 | AC | F | Meth | 67 | IB | N | A | 142 | 142 |
| LCCH01-378/85 | AC | M | Meth | 71 | IA | Y | D | 12 | 23 |
| UD-3 | AC | M | Meth | 44 | IB | Y | D | 60 | 132 |
| UD-7 | AC | M | Meth | 61 | IB | N | A | 84 | 84 |
| UD-18 | AC | F | Meth | 35 | IB | Y | A | 36 | 120 |
| UD-20 | AC | F | Meth | 69 | IIIA | N | A | 108 | 108 |
| UD-24 | AC | F | Meth | 69 | IB | Y | D | 12 | 28 |
| LCCH01-315 | TC | M | Meth, LOH | 54 | IA | N | A | 203 | 288 |
| LCCH01-377/35 | TC | F | Meth, LOH | 35 | IB | N | A | 52 | 52 |
| LCCH01-313 | TC | F | LOH | 57 | IA | N | A | 36 | 36 |
| LCCH01-327 | TC | M | Meth, LOH | 69 | IA | N | A | 66 | 66 |
| LCCH01-320 | TC | F | Meth | 29 | IB | N | A | 157 | 157 |
| LCCH01-323 | TC | F | Meth, LOH | 70 | IA | N | A | 132 | 132 |
| LCCH01-325 | TC | F | Meth | 36 | IA | N | A | 124 | 124 |
| LCCH01-327 | TC | M | Meth | 69 | IA | N | A | 66 | 66 |
| LCCH01-321bis/380 | TC | M | Meth | 48 | IB | N | A | 172 | 172 |
| LCCH01-333 | TC | M | Meth | 71 | IA | N | A | 108 | 108 |
| LCCH01-311 | TC | M | Meth | 50 | IA | N | A | 91 | 91 |
| LCCH01-54 | TC | M | Meth, LOH | 70 | IA | N | A | 47 | 47 |
| UD-9 | TC | M | Meth | 64 | IA | N | A | 72 | 72 |
| UD-10 | TC | M | Meth | 74 | IA | Y | D | 48 | 60 |
| UD-11 | TC | F | Meth | 63 | IA | N | A | 60 | 60 |
| UD-19 | TC | F | Meth | 65 | IB | N | A | 48 | 48 |
| UD-25 | TC | M | Meth | 65 | IA | N | A | 24 | 24 |
| Characteristics | Category | All subjects (N = 47) |
|---|---|---|
| Age at diagnosis (years, median, IQR) | - | 63, 19.75 |
| Gender (n,%) | Female | 26 (55.3%) |
| Male | 21 (44.7%) | |
| Histotype subclassification (n,%) | Typical Carcinoid | 30 (63.8%) |
| Atypical Carcinoid | 17 (36.2%) | |
| T (n,%) | T1 | 31 (67%) |
| T2 | 15 (33%) | |
| N (n,%) | N0 | 41 (89.1%) |
| N1 | 3 (6.5%) | |
| N2 | 2 (4.4%) | |
| M (n,%) | M0 | 47 (100.00%) |
| Tumour stage (n,%) | IA - IB | 42 (89.4%) |
| IIA - IIB - IIIA | 5 (10.6%) |
| Characteristics | Category | UM | M | p-Value * |
|---|---|---|---|---|
| Age at diagnosis (years, median (range)) | - | 63 (30–76) | 64 (29–74) | 0.755 |
| Tumor_size (mm, median (range)) | - | 25 (10–40) | 20 (10–45) | 0.775 |
| Gender | Female | 11 (73%) | 6 (40%) | 0.07 |
| Male | 4 (27%) | 9 (60%) | ||
| T | T1 | 10 (71%) | 11 (73%) | 1.000 |
| T2 | 4 (29%) | 4 (27%) | ||
| N | N0 | 13 (93%) | 15 (100%) | 0.483 |
| N1 | 1 (7%) | 0 (0%) | ||
| M | M0 | 15 (100%) | 15 (100%) | n.a. |
| Tumor stage | IA - IB | 14 (93%) | 15 (100%) | 1.000 |
| IIA-IIB-IIIA | 1 (7%) | 0 (0%) |
| Characteristics | Category | UM | M | p-Value * |
|---|---|---|---|---|
| Age at diagnosis (years, median (range)) | --- | 53 (40–70) | 64 (35–71) | 0.736 |
| Tumor size (mm, median (range)) | --- | 20 (10–48) | 32.5 (20–60) | 0.132 |
| Gender | Female | 5 (56%) | 4 (50%) | 1.000 |
| Male | 4 (44%) | 4 (50%) | ||
| T | T1 | 7 (78%) | 3 (38%) | 0.153 |
| T2 | 2 (22%) | 5 (62%) | ||
| N | N0 | 7 (78%) | 6 (75%) | 0.133 |
| N1 | 2 (22%) | 0 (0%) | ||
| N2 | 0 (0%) | 2 (25%) | ||
| M | M0 | 9 (100%) | 8 (100%) | n.a. |
| Tumor stage | IA-IB | 7 (%) | 6 (%) | 0.893 |
| IIA-IIB-IIIA | 2 (%) | 2 (%) |
| Protein Name (symbol) | Reference |
|---|---|
| NAD(P)H dehydrogenase (quinone 1) (NQO1) | [21], this report |
| Aldo-Keto Reductase Family 1 (AKR1) (AKR1C1) | [22], this report |
| Hemoglobin subunit beta (HBB) | [23,24] |
| Ferritin light chain (FTL) | [24,25] |
| Ferritin heavy chain (FTH) | [24,26,27,28,29] |
| Superoxide dismutase (SOD) | [24,30,31,32,33,34,35] |
| Extracellular Superoxide dismutase (SODE) | [24,36,37] |
| Carbonic Anhydrase 1 (CAH1) | [24,38] |
| Annexin V (ANXA5) | [24,39,40] |
| Peroxiredoxin-2 (PRX2) | [24,41,42,43] |
| Peroxiredoxin-3 (PRX3) | [24,42,44,45] |
| Transthyretin (TTR) | [24,46] |
| Hemopexin (HPX) | [24,47] |
| Transaldolase (TALDO) | [24,48] |
| Heterogenous Nuclear Ribonucleo protein K (HNRPK) | [24,49,50] |
| Histone H4 | [24,51,52,53,54] |
| Heat Shock Protein Hsp90-alpha (HSP90) | [24,55,56,57] |
| Elongation Factor 1-alpha1 (EF1A1) | [24,58] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sparaneo, A.; Fabrizio, F.P.; la Torre, A.; Graziano, P.; Di Maio, M.; Fontana, A.; Bisceglia, M.; Rossi, A.; Pizzolitto, S.; De Maglio, G.; et al. Effects of KEAP1 Silencing on the Regulation of NRF2 Activity in Neuroendocrine Lung Tumors. Int. J. Mol. Sci. 2019, 20, 2531. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20102531
Sparaneo A, Fabrizio FP, la Torre A, Graziano P, Di Maio M, Fontana A, Bisceglia M, Rossi A, Pizzolitto S, De Maglio G, et al. Effects of KEAP1 Silencing on the Regulation of NRF2 Activity in Neuroendocrine Lung Tumors. International Journal of Molecular Sciences. 2019; 20(10):2531. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20102531
Chicago/Turabian StyleSparaneo, Angelo, Federico Pio Fabrizio, Annamaria la Torre, Paolo Graziano, Massimo Di Maio, Andrea Fontana, Michele Bisceglia, Antonio Rossi, Stefano Pizzolitto, Giovanna De Maglio, and et al. 2019. "Effects of KEAP1 Silencing on the Regulation of NRF2 Activity in Neuroendocrine Lung Tumors" International Journal of Molecular Sciences 20, no. 10: 2531. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20102531