Transcriptome Analysis to Shed Light on the Molecular Mechanisms of Early Responses to Cadmium in Roots and Leaves of King Grass (Pennisetum americanum × P. purpureum)
Abstract
:1. Introduction
2. Results and Discussions
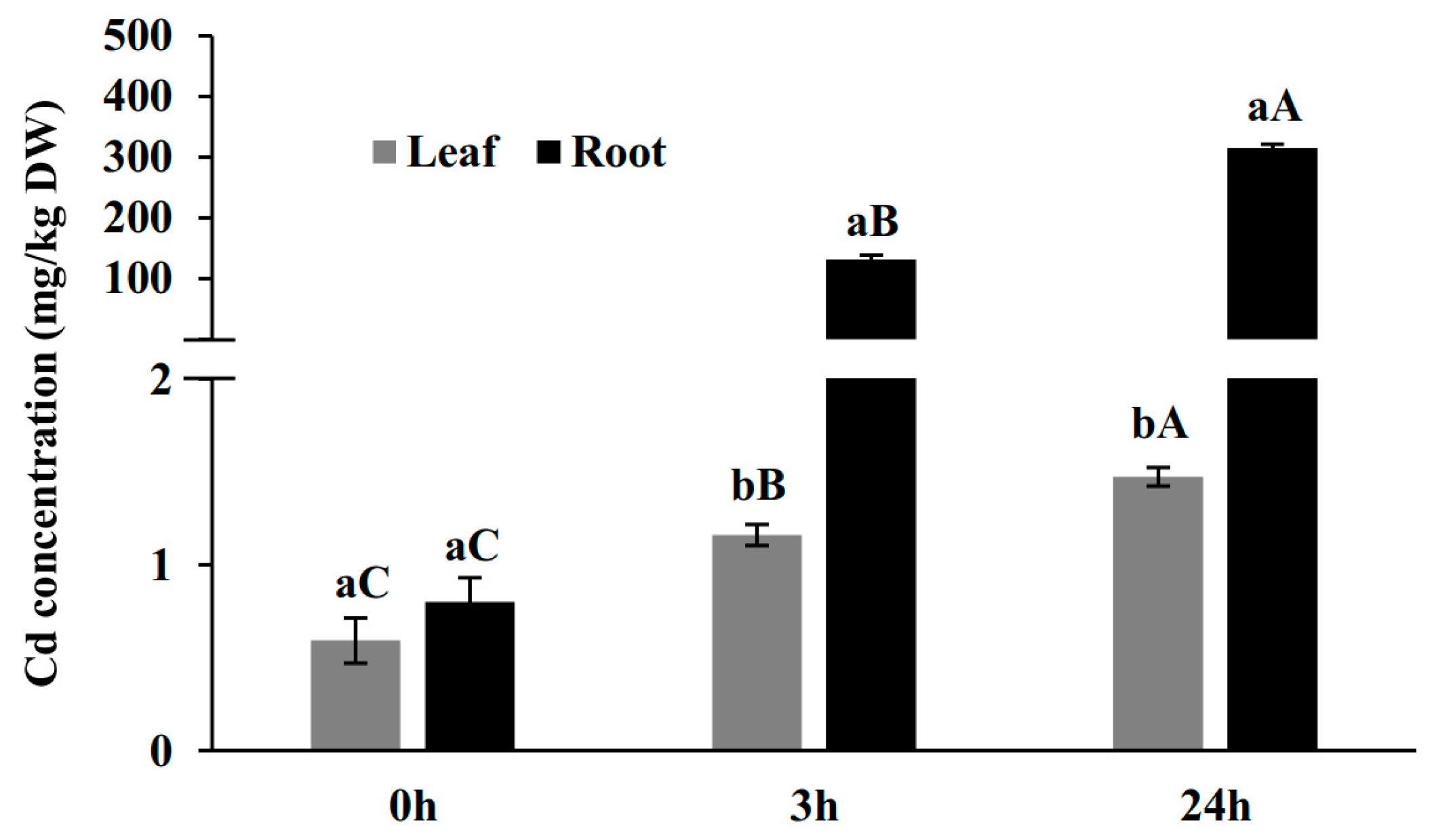
2.1. Cd Concentration in Leaves and Roots in a Time-Course Treatment of 100 µM Cd
2.2. RNA Sequencing and Identification of Differentially Expressed Genes (DEGs)
2.3. RNA Sequencing Validation by qPCR
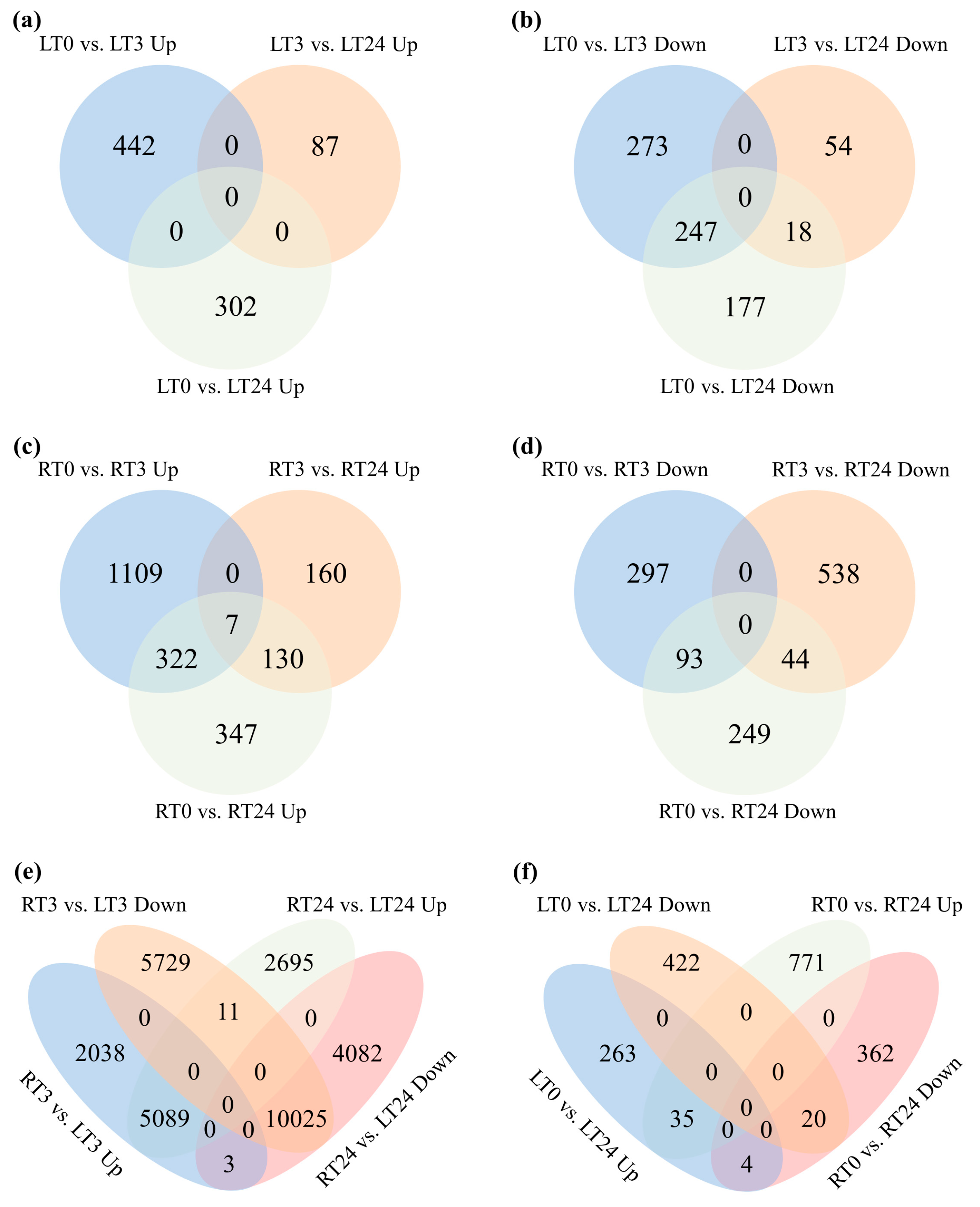
2.4. The Overlaps of DEGs Following CdCl2 Exposure at Different Time Points
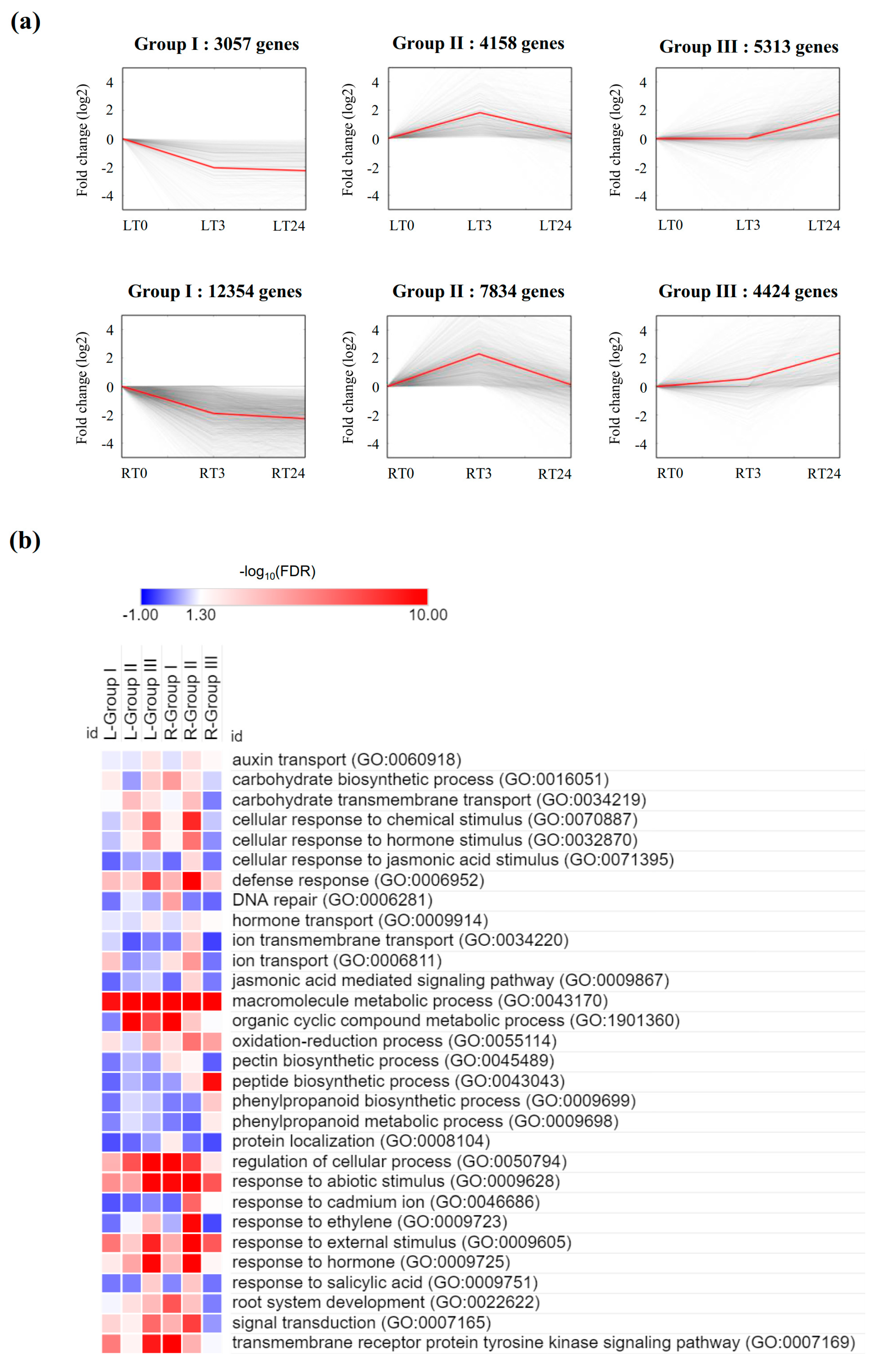
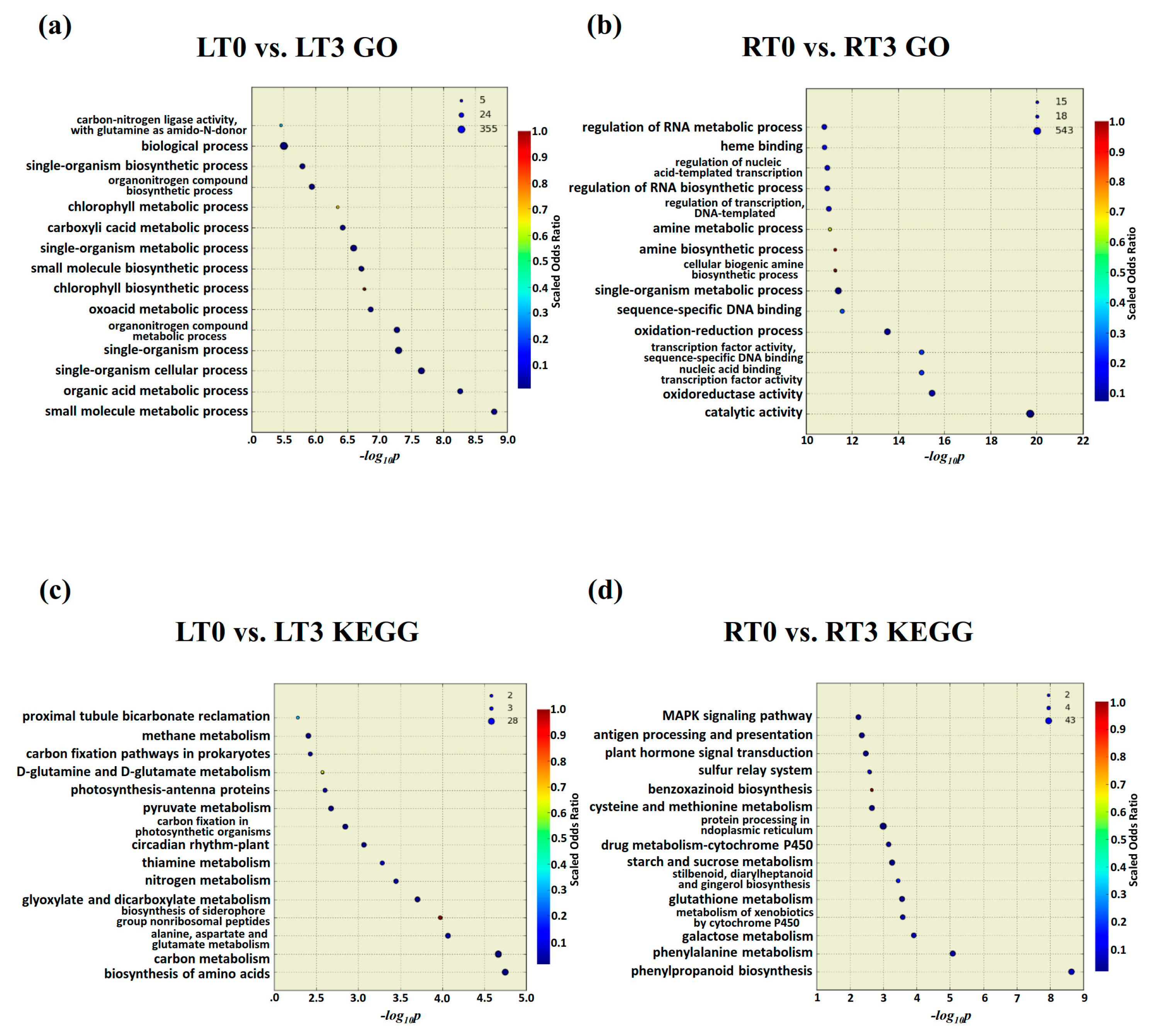
2.5. Gene Expression Trend Analysis, and GO and KEGG Classification
2.6. The Induction of Antioxidant Enzyme-Related Genes May Be Closely Associated with Cd Stress Responses
2.7. Enhanced Cell Wall Biosynthesis Might Play a Vital Role in Cd Stress Tolerance and Cd Ion Accumulations
2.8. Transporter Activities Were Activated in Early Cd Response
2.9. Hormone Signal Transduction Pathway Responded in King Grass
3. Materials and Methods
3.1. Plant Growth and Cd Treatments
3.2. Cd Concentration Measurements in Leaves and Roots
3.3. Determination of ROS Accumulation and the Activities of Antioxidant Enzyme
3.4. RNA Extraction, Sequencing, and De Novo Assembly
3.4.1. RNA Extraction
3.4.2. Library Construction
3.4.3. De Novo Assembly
3.5. Normalization of Gene Expression and DEGs Identification
3.6. Gene Expression Trend Analysis
3.7. Functional Annotation
3.8. Gene Expression Validation by qRT-PCR
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tchounwou, P.B.; Yedjou, C.G.; Patlolla, A.K.; Sutton, D.J. Heavy metal toxicity and the environment. In Molecular, Clinical and Environmental Toxicology; Luch, A., Ed.; Springer: Basel, Switzerland, 2012; Volume 101, pp. 133–164. [Google Scholar]
- Rizwan, M.; Ali, S.; Abbas, T.; Zia-Ur-Rehman, M.; Hannan, F.; Keller, C.; Al-Wabel, M.I.; Yong, S.O. Cadmium minimization in wheat: A critical review. Ecotoxicol. Environ. Saf. 2016, 130, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Rizwan, M.; Ali, S.; Adrees, M.; Ibrahim, M.; Tsang, D.C.W.; Zia-Ur-Rehman, M.; Zahir, Z.A.; Rinklebe, J.; Tack, F.M.G.; Yong, S.O. A critical review on effects, tolerance mechanisms and management of cadmium in vegetables. Chemosphere 2017, 182, 90–105. [Google Scholar] [CrossRef]
- Ali, H.; Khan, E.; Sajad, M.A. Phytoremediation of heavy metals-concepts and applications. Chemosphere 2013, 91, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, K.; Zheng, W.; Zhang, H.; Cao, X.; Lan, Y.; Yang, C.; Li, C. Characterization of early transcriptional responses to cadmium in the root and leaf of Cd-resistant Salix matsudana Koidz. BMC Genom. 2015, 16, 705. [Google Scholar] [CrossRef]
- Zhang, X.F.; Zhang, X.H.; Gao, B.; Li, Z.A.; Xia, H.P.; Li, H.F.; Li, J. Effect of cadmium on growth, photosynthesis, mineral nutrition and metal accumulation of an energy crop, king grass (Pennisetum americanum × P. purpureum). Biomass Bioenerg 2014, 67, 179–187. [Google Scholar] [CrossRef]
- Fan, X.F.; Hou, X.C.; Zhu, Y.; Wu, J.Y. Biomass yield and quality of hybrid pennisetum. Chin. J. Grassl. 2012, 34, 48–52. [Google Scholar]
- Wang, J.; Xin, D.; Hou, X.; Wu, J.; Fan, X.; Li, K.; Zhang, J. Structural properties and hydrolysabilities of chinese pennisetum and hybrid pennisetum: Effect of aqueous ammonia pretreatment. Bioresour. Technol. 2016, 199, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-Cozatl, D.G.; Butko, E.; Springer, F.; Torpey, J.W.; Komives, E.A.; Kehr, J.; Schroeder, J.I. Identification of high levels of phytochelatins, glutathione and cadmium in the phloem sap of Brassica napus. A role for thiol-peptides in the long-distance transport of cadmium and the effect of cadmium on iron translocation. Plant J. 2008, 54, 249–259. [Google Scholar] [CrossRef]
- Guo, W.J.; Meetam, M.; Goldsbrough, P.B. Examining the specific contributions of individual Arabidopsis metallothioneins to copper distribution and metal tolerance. Plant Physiol. 2008, 146, 1697–1706. [Google Scholar] [CrossRef] [PubMed]
- Grennan, A.K. Identification of genes involved in metal transport in plants. Plant Physiol. 2009, 149, 1623–1624. [Google Scholar] [CrossRef] [PubMed]
- Ueno, D.; Iwashita, T.; Zhao, F.J.; Ma, J.F. Characterization of Cd translocation and identification of the Cd form in xylem sap of the Cd-hyperaccumulator Arabidopsis halleri. Plant Cell Physiol. 2008, 49, 540–548. [Google Scholar] [CrossRef]
- Lee, K.; Bae, D.W.; Kim, S.H.; Han, H.J.; Liu, X.; Park, H.C.; Lim, C.O.; Lee, S.Y.; Chung, W.S. Comparative proteomic analysis of the short-term responses of rice roots and leaves to cadmium. J. Plant Physiol. 2010, 167, 161–168. [Google Scholar] [CrossRef]
- He, J.; Li, H.; Luo, J.; Ma, C.; Li, S.; Qu, L.; Gai, Y.; Jiang, X.; Janz, D.; Polle, A. A transcriptomic network underlies microstructural and physiological responses to cadmium in Populus × canescens. Plant Physiol. 2013, 162, 424–439. [Google Scholar] [CrossRef] [PubMed]
- Jozefczak, M.; Keunen, E.; Schat, H.; Bliek, M.; Hernandez, L.E.; Carleer, R.; Remans, T.; Bohler, S.; Vangronsveld, J.; Cuypers, A. Differential response of Arabidopsis leaves and roots to cadmium: Glutathione-related chelating capacity vs. antioxidant capacity. Plant Physiol. Biochem. 2014, 83, 1–9. [Google Scholar] [CrossRef]
- Shamshad, S.; Shahid, M.; Rafiq, M.; Khalid, S.; Dumat, C.; Sabir, M.; Murtaza, B.; Farooq, A.B.U.; Shah, N.S. Effect of organic amendments on cadmium stress to pea: A multivariate comparison of germinating vs. young seedlings and younger vs. older leaves. Ecotoxicol. Environ. Saf. 2018, 151, 91–97. [Google Scholar] [CrossRef]
- Wabnik, K.; Hvidsten, T.R.; Kedzierska, A.; Van Leene, J.; De Jaeger, G.; Beemster, G.T.S.; Komorowski, J.; Kuiper, M.T.R. Gene expression trends and protein features effectively complement each other in gene function prediction. Bioinformatics 2009, 25, 322–330. [Google Scholar] [CrossRef]
- Per, T.S.; Khan, M.I.R.; Anjum, N.A.; Masood, A.; Hussain, S.J.; Khan, N.A. Jasmonates in plants under abiotic stresses: Crosstalk with other phytohormones matters. Environ. Exp. Bot. 2018, 145, 104–120. [Google Scholar] [CrossRef]
- Hsu, Y.T.; Kao, C.H. Toxicity in leaves of rice exposed to cadmium is due to hydrogen peroxide accumulation. Plant Soil 2007, 298, 231–241. [Google Scholar] [CrossRef]
- Shah, K.; Kumar, R.G.; Verma, S.; Dubey, R.S. Effect of cadmium on lipid peroxidation, superoxide anion generation and activities of antioxidant enzymes in growing rice seedlings. Plant Sci. 2001, 161, 1135–1144. [Google Scholar] [CrossRef]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef]
- Li, Z.; Han, X.; Song, X.; Zhang, Y.; Jiang, J.; Han, Q.; Liu, M.; Qiao, G.; Zhuo, R. Overexpressing the Sedum alfredii Cu/Zn superoxide dismutase increased resistance to oxidative stress in transgenic Arabidopsis. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef]
- Alscher, R.G. Biosynthesis and antioxidant function of glutathione in plants. Physiol. Plant. 2010, 77, 457–464. [Google Scholar] [CrossRef]
- Huang, Y.Y.; Shen, C.; Chen, J.X.; He, C.T.; Zhou, Q.; Tan, X.; Yuan, J.G.; Yang, Z.Y. Comparative transcriptome analysis of two Ipomoea aquatica Forsk. Cultivars targeted to explore possible mechanism of genotype-dependent accumulation of cadmium. J. Agric. Food Chem. 2016, 64, 5241–5250. [Google Scholar] [CrossRef]
- Zhou, Q.; Guo, J.J.; He, C.T.; Shen, C.; Huang, Y.Y.; Chen, J.X.; Guo, J.H.; Yuan, J.G.; Yang, Z.Y. Comparative transcriptome analysis between low- and high-cadmium-accumulating genotypes of pakchoi (Brassica chinensis L.) in response to cadmium stress. Environ. Sci. Technol. 2016, 50, 6485–6494. [Google Scholar] [CrossRef] [PubMed]
- Marrs, K.A. The functions and regulation of glutathione S-transferases in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 127–158. [Google Scholar] [CrossRef] [PubMed]
- Clemens, S.; Kim, E.J.; Neumann, D.; Schroeder, J.I. Tolerance to toxic metals by a gene family of phytochelatin synthases from plants and yeast. EMBO J. 1999, 18, 3325–3333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hueckelhoven, R. Cell wall—Associated mechanisms of disease resistance and susceptibility. Ann. Rev. Phytopathol. 2007, 45, 101–127. [Google Scholar] [CrossRef]
- Roberts, K. How the cell wall acquired a cellular context. Plant Physiol. 2001, 125, 127–130. [Google Scholar] [CrossRef]
- Chen, G.; Liu, Y.; Wang, R.; Zhang, J.; Owens, G. Cadmium adsorption by willow root: The role of cell walls and their subfractions. Environ. Sci. Pollut. Res. 2013, 20, 5665–5672. [Google Scholar] [CrossRef]
- DalCorso, G.; Farinati, S.; Furini, A. Regulatory networks of cadmium stress in plants. Plant Signal. Behav. 2010, 5, 663–667. [Google Scholar] [CrossRef]
- Loix, C.; Huybrechts, M.; Vangronsveld, J.; Gielen, M.; Keunen, E.; Cuypers, A. Reciprocal interactions between cadmium-induced cell wall responses and oxidative stress in plants. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Vázquez, S.; Goldsbrough, P.; Carpena, R.O. Assessing the relative contributions of phytochelatins and the cell wall to cadmium resistance in white lupin. Physiol. Plant. 2010, 128, 487–495. [Google Scholar] [CrossRef]
- Ithal, N.; Recknor, J.; Nettleton, D.; Maier, T.; Baum, T.J.; Mitchum, M.G. Developmental transcript profiling of Cyst nematode feeding cells in soybean roots. Mol. Plant-Microbe Interact. 2007, 20, 510–525. [Google Scholar] [CrossRef]
- Magalhaes Silva Moura, J.C.; Valencise Bonine, C.A.; Fernandes Viana, J.d.O.; Dornelas, M.C.; Mazzafera, P. Abiotic and biotic stresses and changes in the lignin content and composition in plants. J. Integr. Plant Biol. 2010, 52, 360–376. [Google Scholar] [CrossRef]
- Parrotta, L.; Guerriero, G.; Sergeant, K.; Cal, G.; Hausman, J.-F. Target or barrier? The cell wall of early- and later-diverging plants vs. cadmium toxicity: Differences in the response mechanisms. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef]
- Herbette, S.; Taconnat, L.; Hugouvieux, V.; Piette, L.; Magniette, M.L.M.; Cuine, S.; Auroy, P.; Richaud, P.; Forestier, C.; Bourguignon, J.; et al. Genome-wide transcriptome profiling of the early cadmium response of Arabidopsis roots and shoots. Biochimie 2006, 88, 1751–1765. [Google Scholar] [CrossRef] [PubMed]
- Kovacik, J.; Backor, M.; Kadukova, J. Physiological responses of matricaria chamomilla to cadmium and copper excess. Environ. Toxicol. 2008, 23, 123–130. [Google Scholar] [CrossRef]
- Rui, H.; Chen, C.; Zhang, X.; Shen, Z.; Zhang, F. Cd-induced oxidative stress and lignification in the roots of two Vicia sativa L. varieties with different Cd tolerances. J. Hazard. Mater. 2016, 301, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Roth, U.; von Roepenack-Lahaye, E.; Clemens, S. Proteome changes in Arabidopsis thaliana roots upon exposure to Cd2+. J. Exp. Bot. 2006, 57, 4003–4013. [Google Scholar] [CrossRef] [PubMed]
- Guerinot, M.L. The ZIP family of metal transporters. Biochim. Biophys. Acta 2000, 1465, 190–198. [Google Scholar] [CrossRef] [Green Version]
- Vert, G.; Grotz, N.; Dedaldechamp, F.; Gaymard, F.; Guerinot, M.L.; Briat, J.-F.; Curie, C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 2002, 14, 1223–1233. [Google Scholar] [CrossRef]
- Connolly, E.L.; Fett, J.P.; Guerinot, M.L. Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 2002, 14, 1347–1357. [Google Scholar] [CrossRef]
- Lee, S.; An, G. Over-expression of OsIRT1 leads to increased iron and zinc accumulations in rice. Plant Cell Environ. 2009, 32, 408–416. [Google Scholar] [CrossRef]
- Lombi, E.; Tearall, K.L.; Howarth, J.R.; Zhao, F.-J.; Hawkesford, M.J.; McGrath, S.P. Influence of iron status on cadmium and zinc uptake by different ecotypes of the hyperaccumulator Thlaspi caerulescens. Plant Physiol. 2002, 128, 1359–1367. [Google Scholar] [CrossRef]
- Chi, S.; Qin, Y.; Xu, W.; Chai, Y.; Feng, D.; Li, Y.; Li, T.; Yang, M.; He, Z. Differences of Cd uptake and expression of OAS and IRT genes in two varieties of ryegrasses. Environ. Sci. Pollut. Res. Int. 2018. [Google Scholar] [CrossRef]
- Klein, I.; Sarkadi, B.; Váradi, A. An inventory of the human ABC proteins. Biochim. Biophys. Acta 2000, 1461, 237–262. [Google Scholar] [CrossRef]
- Martinoia, E.; Klein, M.; Geisler, M.; Bovet, L.; Forestier, C.; Kolukisaoglu, U.; Muller-Rober, B.; Schulz, B. Multifunctionality of plant ABC transporters--more than just detoxifiers. Planta 2002, 214, 345–355. [Google Scholar] [CrossRef]
- Ortiz, D.F.; Ruscitti, T.; McCue, K.F.; Ow, D.W. Transport of metal-binding peptides by HMT1, a fission yeast ABC-type vacuolar membrane protein. J. Biol. Chem. 1995, 270, 4721–4728. [Google Scholar] [CrossRef]
- Kim, D.Y.; Bovet, L.; Maeshima, M.; Martinoia, E.; Lee, Y. The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 2007, 50, 207–218. [Google Scholar] [CrossRef]
- Wojas, S.; Hennig, J.; Plaza, S.; Geisler, M.; Siemianowski, O.; Sklodowska, A.; Ruszczynska, A.; Bulska, E.; Antosiewicz, D.M. Ectopic expression of Arabidopsis ABC transporter MRP7 modifies cadmium root-to-shoot transport and accumulation. Environ. Pollut. 2009, 157, 2781–2789. [Google Scholar] [CrossRef]
- Lu, Y.P.; Li, Z.S.; Rea, P.A. AtMRP1 gene of Arabidopsis encodes a glutathione S-conjugate pump: Isolation and functional definition of a plant ATP-binding cassette transporter gene. Proc. Natl. Acad. Sci. USA 1997, 94, 8243–8248. [Google Scholar] [CrossRef]
- Migocka, M.; Papierniak, A.; Kosatka, E.; Kłobus, G. Comparative study of the active cadmium efflux systems operating at the plasma membrane and tonoplast of cucumber root cells. J. Exp. Bot. 2011, 62, 4903–4916. [Google Scholar] [CrossRef]
- Yan, J.; Wang, P.; Wang, P.; Yang, M.; Lian, X.; Tang, Z.; Huang, C.-F.; Salt, D.E.; Zhao, F.J. A loss-of-function allele of OsHMA3 associated with high cadmium accumulation in shoots and grain of japonica rice cultivars. Plant Cell Environ. 2016, 39, 1941–1954. [Google Scholar] [CrossRef]
- Ueno, D.; Yamaji, N.; Kono, I.; Huang, C.F.; Ando, T.; Yano, M.; Ma, J.F. Gene limiting cadmium accumulation in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 16500–16505. [Google Scholar] [CrossRef] [Green Version]
- Baxter, I.; Tchieu, J.; Sussman, M.R.; Boutry, M.; Palmgren, M.G.; Gribskov, M.; Harper, J.F.; Axelsen, K.B. Genomic comparison of P-type ATPase ion pumps in Arabidopsis and rice. Plant Physiol. 2003, 132, 618–628. [Google Scholar] [CrossRef]
- Williams, L.E.; Mills, R.F. P(1b)-ATPases--an ancient family of transition metal pumps with diverse functions in plants. Trends Plant Sci. 2005, 10, 491–502. [Google Scholar] [CrossRef]
- Hussain, D.; Haydon, M.J.; Wang, Y.; Wong, E.; Sherson, S.M.; Young, J.; Camakaris, J.; Harper, J.F.; Cobbett, C.S. P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell 2004, 16, 1327–1339. [Google Scholar] [CrossRef]
- Mills, R.F.; Valdes, B.; Duke, M.; Peaston, K.A.; Lahner, B.; Salt, D.E.; Williams, L.E. Functional significance of AtHMA4 C-terminal domain in planta. PLoS ONE 2010, 5. [Google Scholar] [CrossRef]
- Feng, J.J.; Jia, W.T.; Lv, S.L.; Bao, H.; Miao, F.F.; Zhang, X.; Wang, J.H.; Li, J.H.; Li, D.S.; Zhu, C.; et al. Comparative transcriptome combined with morpho-physiological analyses revealed key factors for differential cadmium accumulation in two contrasting sweet sorghum genotypes. Plant Biotechnol. J. 2018, 16, 558–571. [Google Scholar] [CrossRef]
- Verbruggen, N.; Hermans, C.; Schat, H. Molecular mechanisms of metal hyperaccumulation in plants. New Phytol. 2009, 181, 759–776. [Google Scholar] [CrossRef]
- Chmielowska-Bak, J.; Gzyl, J.; Rucińska-Sobkowiak, R.; Arasimowicz-Jelonek, M.; Deckert, J. The new insights into cadmium sensing. Front. Plant Sci. 2014, 5, 245. [Google Scholar]
- Ku, Y.S.; Sintaha, M.; Cheung, M.Y.; Lam, H.M. Plant hormone signaling crosstalks between biotic and abiotic stress responses. Int. J. Mol. Sci. 2014, 19, 3206. [Google Scholar] [CrossRef]
- Noriega, G.; Santa Cruz, D.; Batlle, A.; Tomaro, M.; Balestrasse, K. Heme oxygenase is involved in the protection exerted by jasmonic acid against cadmium stress in soybean roots. J. Plant Growth Regul. 2012, 31, 79–89. [Google Scholar] [CrossRef]
- Rodriguez-Serrano, M.; Romero-Puertas, M.C.; Zabalza, A.; Corpas, F.J.; Gomez, M.; Del Rio, L.A.; Sandalio, L.M. Cadmium effect on oxidative metabolism of pea (Pisum sativum L.) roots. Imaging of reactive oxygen species and nitric oxide accumulation in vivo. Plant Cell Environ. 2006, 29, 1532–1544. [Google Scholar] [CrossRef]
- Maksymiec, W.; Wianowska, D.; Dawidowicz, A.L.; Radkiewicz, S.; Mardarowicz, M.; Krupa, Z. The level of jasmonic acid in Arabidopsis thaliana and Phaseolus coccineus plants under heavy metal stress. J. Plant Physiol. 2005, 162, 1338–1346. [Google Scholar] [CrossRef]
- Maksymiec, W.; Krupa, Z. Jasmonic acid and heavy metals in Arabidopsis plants—A similar physiological response to both stressors? J. Plant Physiol. 2002, 159, 509–515. [Google Scholar] [CrossRef]
- Dutilleul, C.; Garmier, M.; Noctor, G.; Mathieu, C.; Chetrit, P.; Foyer, C.H.; Paepe, R.D. Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell 2003, 15, 1212–1226. [Google Scholar] [CrossRef]
- Chance, B.; Maehly, A.C. Assay of catalases and peroxidases. Methods Enzymol. 1955, 2, 764–775. [Google Scholar]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nature Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Li, W.Z.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. Rsem: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Nikolayeva, O.; Robinson, M.D. Edger for differential RNA-seq and CHIP-seq analysis: An application to stem cell biology. Methods Mol. Biol. 2014, 1150, 45–79. [Google Scholar] [PubMed]
- Reddy, P.S.; Reddy, D.S.; Sharma, K.K.; Bhatnagar-Mathur, P.; Vadez, V. Cloning and validation of reference genes for normalization of gene expression studies in pearl millet [Pennisetum glaucum (L.) R. Br.] by quantitative real-time PCR. Plant Gene 2015, 1, 35–42. [Google Scholar] [CrossRef]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, J.; Xia, B.; Meng, Y.; Yang, Z.; Pan, L.; Zhou, M.; Zhang, X. Transcriptome Analysis to Shed Light on the Molecular Mechanisms of Early Responses to Cadmium in Roots and Leaves of King Grass (Pennisetum americanum × P. purpureum). Int. J. Mol. Sci. 2019, 20, 2532. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20102532
Zhao J, Xia B, Meng Y, Yang Z, Pan L, Zhou M, Zhang X. Transcriptome Analysis to Shed Light on the Molecular Mechanisms of Early Responses to Cadmium in Roots and Leaves of King Grass (Pennisetum americanum × P. purpureum). International Journal of Molecular Sciences. 2019; 20(10):2532. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20102532
Chicago/Turabian StyleZhao, Junming, Bo Xia, Yu Meng, Zhongfu Yang, Ling Pan, Man Zhou, and Xinquan Zhang. 2019. "Transcriptome Analysis to Shed Light on the Molecular Mechanisms of Early Responses to Cadmium in Roots and Leaves of King Grass (Pennisetum americanum × P. purpureum)" International Journal of Molecular Sciences 20, no. 10: 2532. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20102532



