A Vector-Based Method to Analyze the Topography of Glial Networks
Abstract
:1. Introduction
2. Results
2.1. Astrocyte Networks
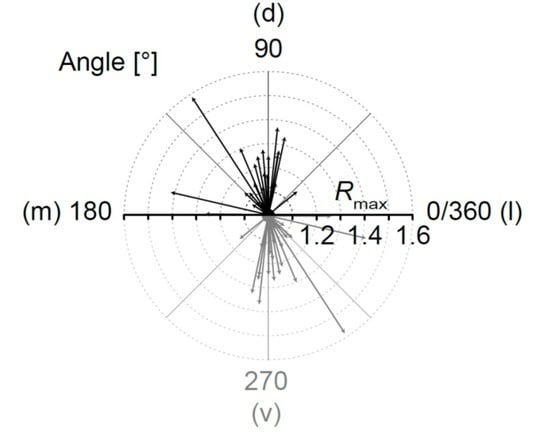
2.2. Analysis of Network Topography
2.3. Meta-Analysis of “Vector Means”
2.4. Performance of Approaches
3. Discussion
3.1. Intensity-Based Cell Detection Method
3.2. Comparison of Approaches
3.3. Astrocyte Maturation and Network Topography
3.4. Tracer Coupling versus Electrical Coupling
3.5. Conclusions
4. Materials and Methods
4.1. Preparation of Acute Brainstem Slices
4.2. Tracer Loading
4.3. Visualization of Coupled Cells
4.4. Confocal Microscopy
4.5. Reconstruction of Gap Junction Networks
4.6. Analysis of Network Topography
4.6.1. Manual “YX Ratio”
4.6.2. Automated “YX Ratio”
4.6.3. “Intensity + Coordinates”
4.6.4. “Intensity Profiles”
4.6.5. “Vector Sum”
4.6.6. “Vector Means”
4.6.7. Meta-Analysis
4.6.8. Generation of Artificial Networks In Silico
4.7. Statistics
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ACSF | Artificial cerebrospinal fluid |
| BSA | Bovine serum albumin |
| Ctx | Cortex |
| Cx | Connexin |
| FWHM | Full-width at half-maximum |
| GJ | Gap junction |
| HC | Hippocampus |
| IC | Inferior colliculus |
| LSO | Lateral superior olive |
| Nb+ | Neurobiotin-positive |
| NGS | Normal goat serum |
| nPA | Nonpassive astrocyte |
| PA | Passive astrocyte |
| PBS | Phosphate buffered solution |
| PFA | Paraformaldehyde |
| R | Ratio |
| R2 | Regression coefficient |
| RT | Room temperature |
| ROI | Region of interest |
| SR101 | Sulforhodamine 101 |
| Th | Thalamus |
References
- Giaume, C.; Theis, M. Pharmacological and genetic approaches to study connexin-mediated channels in glial cells of the central nervous system. Brain Res. Rev. 2010, 63, 160–176. [Google Scholar] [CrossRef] [PubMed]
- Goodenough, D.A.; Paul, D.L. Gap junctions. Cold Spring Harb. Perspect. Biol. 2009, 1, a002576. [Google Scholar] [CrossRef] [PubMed]
- Maglione, M.; Tress, O.; Haas, B.; Karram, K.; Trotter, J.; Willecke, K.; Kettenmann, H. Oligodendrocytes in mouse corpus callosum are coupled via gap junction channels formed by connexin47 and connexin32. Glia 2010, 58, 1104–1117. [Google Scholar] [CrossRef] [PubMed]
- Augustin, V.; Bold, C.; Wadle, S.L.; Langer, J.; Jabs, R.; Philippot, C.; Weingarten, D.J.; Rose, C.R.; Steinhauser, C.; Stephan, J. Functional anisotropic panglial networks in the lateral superior olive. Glia 2016, 64, 1892–1911. [Google Scholar] [CrossRef]
- Wadle, S.L.; Augustin, V.; Langer, J.; Jabs, R.; Philippot, C.; Weingarten, D.J.; Rose, C.R.; Steinhauser, C.; Stephan, J. Anisotropic Panglial Coupling Reflects Tonotopic Organization in the Inferior Colliculus. Front. Cell. Neurosci. 2018, 12, 431. [Google Scholar] [CrossRef] [PubMed]
- Moshrefi-Ravasdjani, B.; Hammel, E.L.; Kafitz, K.W.; Rose, C.R. Astrocyte Sodium Signalling and Panglial Spread of Sodium Signals in Brain White Matter. Neurochem. Res. 2017, 42, 2505–2518. [Google Scholar] [CrossRef] [PubMed]
- Claus, L.; Philippot, C.; Griemsmann, S.; Timmermann, A.; Jabs, R.; Henneberger, C.; Kettenmann, H.; Steinhauser, C. Barreloid Borders and Neuronal Activity Shape Panglial Gap Junction-Coupled Networks in the Mouse Thalamus. Cereb. Cortex 2018, 28, 213–222. [Google Scholar] [CrossRef] [PubMed]
- Griemsmann, S.; Hoft, S.P.; Bedner, P.; Zhang, J.; von Staden, E.; Beinhauer, A.; Degen, J.; Dublin, P.; Cope, D.W.; Richter, N.; et al. Characterization of Panglial Gap Junction Networks in the Thalamus, Neocortex, and Hippocampus Reveals a Unique Population of Glial Cells. Cereb. Cortex 2015, 25, 3420–3433. [Google Scholar] [CrossRef] [PubMed]
- Binmoller, F.J.; Muller, C.M. Postnatal development of dye-coupling among astrocytes in rat visual cortex. Glia 1992, 6, 127–137. [Google Scholar] [CrossRef]
- Houades, V.; Rouach, N.; Ezan, P.; Kirchhoff, F.; Koulakoff, A.; Giaume, C. Shapes of astrocyte networks in the juvenile brain. Neuron Glia Biol. 2006, 2, 3–14. [Google Scholar] [CrossRef] [Green Version]
- Houades, V.; Koulakoff, A.; Ezan, P.; Seif, I.; Giaume, C. Gap junction-mediated astrocytic networks in the mouse barrel cortex. J. Neurosci. 2008, 28, 5207–5217. [Google Scholar] [CrossRef] [PubMed]
- Condamine, S.; Lavoie, R.; Verdier, D.; Kolta, A. Functional rhythmogenic domains defined by astrocytic networks in the trigeminal main sensory nucleus. Glia 2018, 66, 311–326. [Google Scholar] [CrossRef] [PubMed]
- Condamine, S.; Verdier, D.; Kolta, A. Analyzing the Size, Shape, and Directionality of Networks of Coupled Astrocytes. J. Vis. Exp. 2018, 140, e58116. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Minge, D.; Griemsmann, S.; Herde, M.K.; Steinhauser, C.; Henneberger, C. Spatial properties of astrocyte gap junction coupling in the rat hippocampus. Philos. Trans. R Soc. B Biol. Sci. 2014, 369, 20130600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghezali, G.; Calvo, C.F.; Pillet, L.E.; Llense, F.; Ezan, P.; Pannasch, U.; Bemelmans, A.P.; Etienne Manneville, S.; Rouach, N. Connexin 30 controls astroglial polarization during postnatal brain development. Development 2018, 145. [Google Scholar] [CrossRef] [PubMed]
- Kandler, K.; Clause, A.; Noh, J. Tonotopic reorganization of developing auditory brainstem circuits. Nat. Neurosci. 2009, 12, 711–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stephan, J.; Friauf, E. Functional analysis of the inhibitory neurotransmitter transporters GlyT1, GAT-1, and GAT-3 in astrocytes of the lateral superior olive. Glia 2014, 62, 1992–2003. [Google Scholar] [CrossRef] [PubMed]
- Kafitz, K.W.; Meier, S.D.; Stephan, J.; Rose, C.R. Developmental profile and properties of sulforhodamine 101--Labeled glial cells in acute brain slices of rat hippocampus. J. Neurosci. Methods 2008, 169, 84–92. [Google Scholar] [CrossRef]
- Ghirardini, E.; Wadle, S.L.; Augustin, V.; Becker, J.; Brill, S.; Hammerich, J.; Seifert, G.; Stephan, J. Expression of functional inhibitory neurotransmitter transporters GlyT1, GAT-1, and GAT-3 by astrocytes of inferior colliculus and hippocampus. Mol. Brain 2018, 11, 4. [Google Scholar] [CrossRef]
- Nimmerjahn, A.; Helmchen, F. In vivo labeling of cortical astrocytes with sulforhodamine 101 (SR101). Cold Spring Harb. Protoc. 2012, 2012, 326–334. [Google Scholar] [CrossRef]
- Yagi, T.; Terada, N.; Baba, T.; Ohno, S. Localization of endogenous biotin-containing proteins in mouse Bergmann glial cells. Histochem. J. 2002, 34, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Bixel, M.G.; Hamprecht, B. Immunocytochemical localization of beta-methylcrotonyl-CoA carboxylase in astroglial cells and neurons in culture. J. Neurochem. 2000, 74, 1059–1067. [Google Scholar] [CrossRef] [PubMed]
- Livnah, O.; Bayer, E.A.; Wilchek, M.; Sussman, J.L. Three-dimensional structures of avidin and the avidin-biotin complex. Proc. Natl. Acad. Sci. USA 1993, 90, 5076–5080. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Schools, G.P.; Kimelberg, H.K. Development of GLAST(+) astrocytes and NG2(+) glia in rat hippocampus CA1: Mature astrocytes are electrophysiologically passive. J. Neurophysiol. 2006, 95, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Schools, G.P.; Zhou, M.; Kimelberg, H.K. Development of gap junctions in hippocampal astrocytes: Evidence that whole cell electrophysiological phenotype is an intrinsic property of the individual cell. J. Neurophysiol. 2006, 96, 1383–1392. [Google Scholar] [CrossRef]
- Wallraff, A.; Odermatt, B.; Willecke, K.; Steinhauser, C. Distinct types of astroglial cells in the hippocampus differ in gap junction coupling. Glia 2004, 48, 36–43. [Google Scholar] [CrossRef]
- Muller, J.; Reyes-Haro, D.; Pivneva, T.; Nolte, C.; Schaette, R.; Lubke, J.; Kettenmann, H. The principal neurons of the medial nucleus of the trapezoid body and NG2(+) glial cells receive coordinated excitatory synaptic input. J. Gen. Physiol. 2009, 134, 115–127. [Google Scholar] [CrossRef]
- Xu, G.; Wang, W.; Zhou, M. Spatial organization of NG2 glial cells and astrocytes in rat hippocampal CA1 region. Hippocampus 2014, 24, 383–395. [Google Scholar] [CrossRef]
- Roux, L.; Benchenane, K.; Rothstein, J.D.; Bonvento, G.; Giaume, C. Plasticity of astroglial networks in olfactory glomeruli. Proc. Natl. Acad. Sci. USA 2011, 108, 18442–18446. [Google Scholar] [CrossRef] [Green Version]
- Ma, B.; Buckalew, R.; Du, Y.; Kiyoshi, C.M.; Alford, C.C.; Wang, W.; McTigue, D.M.; Enyeart, J.J.; Terman, D.; Zhou, M. Gap junction coupling confers isopotentiality on astrocyte syncytium. Glia 2016, 64, 214–226. [Google Scholar] [CrossRef]
- Huang, M.; Du, Y.; Kiyoshi, C.; Wu, X.; Askwith, C.; McTigue, D.; Zhou, M. Syncytial Isopotentiality: An Electrical Feature of Spinal Cord Astrocyte Networks. Neuroglia 2018, 1, 18. [Google Scholar] [CrossRef]
- Kiyoshi, C.M.; Du, Y.; Zhong, S.; Wang, W.; Taylor, A.T.; Xiong, B.; Ma, B.; Terman, D.; Zhou, M. Syncytial isopotentiality: A system-wide electrical feature of astrocytic networks in the brain. Glia 2018, 66, 2756–2769. [Google Scholar] [CrossRef] [PubMed]
- Langer, J.; Stephan, J.; Theis, M.; Rose, C.R. Gap junctions mediate intercellular spread of sodium between hippocampal astrocytes in situ. Glia 2012, 60, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Pusch, M.; Neher, E. Rates of diffusional exchange between small cells and a measuring patch pipette. Pflug. Arch. 1988, 411, 204–211. [Google Scholar] [CrossRef]
- Friauf, E.; Aragon, C.; Lohrke, S.; Westenfelder, B.; Zafra, F. Developmental expression of the glycine transporter GLYT2 in the auditory system of rats suggests involvement in synapse maturation. J. Comp. Neurol. 1999, 412, 17–37. [Google Scholar] [CrossRef]
- Hirtz, J.J.; Braun, N.; Griesemer, D.; Hannes, C.; Janz, K.; Lohrke, S.; Muller, B.; Friauf, E. Synaptic refinement of an inhibitory topographic map in the auditory brainstem requires functional Cav1.3 calcium channels. J. Neurosci. 2012, 32, 14602–14616. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]






| Study | Approach | nPA (classes) | PA (classes) | p | ||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 1 | 2 | 3 | |||
| This study (LSO) | YX ratio (manual) | 79% | 14% | 7% | 40% | 50% | 10% | 1.6096 × 10−14 |
| YX ratio (automatic) | 64% | 36% | 0% | 50% | 30% | 20% | 3.4229 × 10−6 | |
| Intensity + coordinates | 93% | 0% | 7% | 60% | 20% | 20% | 9.0177 × 10−11 | |
| Intensity profiles | 57% | 43% | 0% | 30% | 70% | 0% | 3.1601 × 10−9 | |
| Vector means | 79% | 21% | 0% | 30% | 60% | 10% | 2.3325 × 10−25 | |
| Previous studies | ||||||||
| [4] (LSO) | YX ratio (manual) | 58% | 33% | 8% | 39% | 39% | 22% | 6.7850 × 10−5 |
| [5] (IC) | YX ratio (manual) | 71% | 21% | 7% | 43% | 43% | 13% | 1.0375 × 10−7 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eitelmann, S.; Hirtz, J.J.; Stephan, J. A Vector-Based Method to Analyze the Topography of Glial Networks. Int. J. Mol. Sci. 2019, 20, 2821. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20112821
Eitelmann S, Hirtz JJ, Stephan J. A Vector-Based Method to Analyze the Topography of Glial Networks. International Journal of Molecular Sciences. 2019; 20(11):2821. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20112821
Chicago/Turabian StyleEitelmann, Sara, Jan J. Hirtz, and Jonathan Stephan. 2019. "A Vector-Based Method to Analyze the Topography of Glial Networks" International Journal of Molecular Sciences 20, no. 11: 2821. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20112821