Translation Control of HAC1 by Regulation of Splicing in Saccharomyces cerevisiae
Abstract
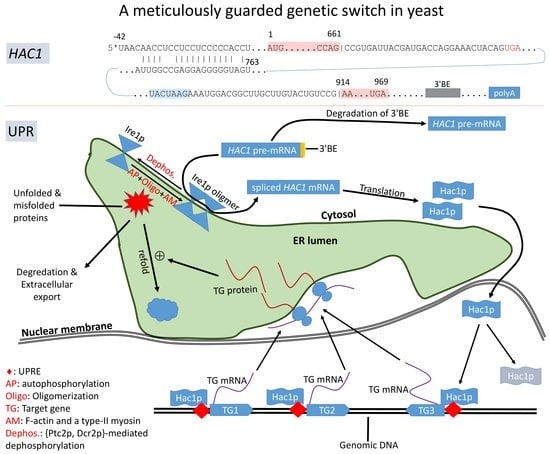
:1. Introduction
2. HAC1 and Its Translation Control via Ire1p-Mediated Splicing
2.1. Intron-Mediated Translation Control of HAC1 mRNA
2.2. Ire1p-Mediated Splicing of HAC1 mRNA
2.3. Hac1p Triggers UPR and Is Autoregulated
3. Leaky Splicing of HAC1 mRNA in Non-UPR Yeast Cells
4. Leaky Translation of HAC1u
5. Ire1p Domain Structure and Its Splicing Activity
6. How Is Ire1p Activity Regulated in Response to Unfolded/Misfolded Proteins in Yeast?
7. Conservation and Diversification of Ire1p-Mediated UPR Signaling
7.1. Conservation and Diversification of the Ire1p-Hac1p Pathway
7.2. Conservation and Diversity of Ire1p-Mediated Splicing
7.3. Diversity in Translation Control
8. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Zid, B.M.; Rogers, A.N.; Katewa, S.D.; Vargas, M.A.; Kolipinski, M.C.; Lu, T.A.; Benzer, S.; Kapahi, P. 4e-bp extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell 2009, 139, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, W.V.; Zhou, K.; Butler, T.K.; Doudna, J.A. Cap-independent translation is required for starvation-induced differentiation in yeast. Science 2007, 317, 1224–1227. [Google Scholar] [CrossRef] [PubMed]
- Fauchon, M.; Lagniel, G.; Aude, J.C.; Lombardia, L.; Soularue, P.; Petat, C.; Marguerie, G.; Sentenac, A.; Werner, M.; Labarre, J. Sulfur sparing in the yeast proteome in response to sulfur demand. Mol. Cell 2002, 9, 713–723. [Google Scholar] [CrossRef]
- Gebauer, F.; Preiss, T.; Hentze, M.W. From cis-regulatory elements to complex RNPs and back. Cold Spring Harb. Perspect. Biol. 2012, 4, a012245. [Google Scholar] [CrossRef] [PubMed]
- Roux, P.P.; Topisirovic, I. Regulation of mRNA translation by signaling pathways. Cold Spring Harb. Perspect. Biol. 2012, 4, a012252. [Google Scholar] [CrossRef] [PubMed]
- De Melo Neto, O.P.; Standart, N.; Martins de Sa, C. Autoregulation of poly(a)-binding protein synthesis in vitro. Nucleic. Acids Res. 1995, 23, 2198–2205. [Google Scholar] [CrossRef]
- Wu, J.; Bag, J. Negative control of the poly(a)-binding protein mRNA translation is mediated by the adenine-rich region of its 5’-untranslated region. J. Biol. Chem. 1998, 273, 34535–34542. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; MacKay, V.; Yao, X.; Wu, J.; Miura, F.; Ito, T.; Morris, D.R. Translation initiation: A regulatory role for poly(a) tracts in front of the AUG codon in Saccharomyces cerevisiae. Genetics 2011, 189, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Craigen, W.J.; Caskey, C.T. Expression of peptide chain release factor 2 requires high-efficiency frameshift. Nature 1986, 322, 273–275. [Google Scholar] [CrossRef]
- Craigen, W.J.; Cook, R.G.; Tate, W.P.; Caskey, C.T. Bacterial peptide chain release factors: Conserved primary structure and possible frameshift regulation of release factor 2. Proc. Natl. Acad. Sci. USA 1985, 82, 3616–3620. [Google Scholar] [CrossRef] [PubMed]
- Xia, X. Bioinformatics and translation termination in bacteria. In Bioinformatics and the Cell: Modern Computational Approaches in Genomics, Proteomics and Transcriptomics; Springer: Cham, Switzerland, 2018; pp. 239–254. [Google Scholar]
- Wei, Y.; Wang, J.; Xia, X. Coevolution between stop codon usage and release factors in bacterial species. Mol. Biol. Evol. 2016, 33, 2357–2367. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Xia, X. The role of +4u as an extended translation termination signal in bacteria. Genetics 2017, 205, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Nojima, H.; Leem, S.H.; Araki, H.; Sakai, A.; Nakashima, N.; Kanaoka, Y.; Ono, Y. Hac1: A novel yeast bZIP protein binding to the CRE motif is a multicopy suppressor for cdc10 mutant of Schizosaccharomyces pombe. Nucleic. Acids Res. 1994, 22, 5279–5288. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Kawahara, T.; Yoshida, H.; Yanagi, H.; Yura, T. Signalling from endoplasmic reticulum to nucleus: Transcription factor with a basic-leucine zipper motif is required for the unfolded protein-response pathway. Genes Cells 1996, 1, 803–817. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.S.; Walter, P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 1996, 87, 391–404. [Google Scholar] [CrossRef]
- Chapman, R.E.; Walter, P. Translational attenuation mediated by an mRNA intron. Curr. Biol. 1997, 7, 850–859. [Google Scholar] [CrossRef] [Green Version]
- Nikawa, J.; Akiyoshi, M.; Hirata, S.; Fukuda, T. Saccharomyces cerevisiae IRE2/HAC1 is involved in IRE1-mediated KAR2 expression. Nucleic. Acids Res. 1996, 24, 4222–4226. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Ogawa, N.; Kawahara, T.; Yanagi, H.; Yura, T. Palindrome with spacer of one nucleotide is characteristic of the cis-acting unfolded protein response element in Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 9912–9920. [Google Scholar] [CrossRef] [PubMed]
- Sidrauski, C.; Cox, J.S.; Walter, P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell 1996, 87, 405–413. [Google Scholar] [CrossRef]
- Sidrauski, C.; Walter, P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell 1997, 90, 1031–1039. [Google Scholar] [CrossRef]
- Gonzalez, T.N.; Sidrauski, C.; Dorfler, S.; Walter, P. Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. Embo. J. 1999, 18, 3119–3132. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, R.J. Stress signaling from the lumen of the endoplasmic reticulum: Coordination of gene transcriptional and translational controls. Genes Dev. 1999, 13, 1211–1233. [Google Scholar] [CrossRef] [PubMed]
- Welihinda, A.A.; Tirasophon, W.; Kaufman, R.J. The cellular response to protein misfolding in the endoplasmic reticulum. Gene. Expr. 1999, 7, 293–300. [Google Scholar] [PubMed]
- Welihinda, A.A.; Tirasophon, W.; Kaufman, R.J. The transcriptional co-activator ada5 is required for HAC1 mRNA processing in vivo. J. Biol. Chem. 2000, 275, 3377–3381. [Google Scholar] [CrossRef]
- Aragon, T.; van Anken, E.; Pincus, D.; Serafimova, I.M.; Korennykh, A.V.; Rubio, C.A.; Walter, P. Messenger RNA targeting to endoplasmic reticulum stress signalling sites. Nature 2009, 457, 736–740. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, T.; Yanagi, H.; Yura, T.; Mori, K. Unconventional splicing of HAC1/ern4 mRNA required for the unfolded protein response. Sequence-specific and non-sequential cleavage of the splice sites. J. Biol. Chem. 1998, 273, 1802–1807. [Google Scholar] [CrossRef]
- Mori, K.; Ogawa, N.; Kawahara, T.; Yanagi, H.; Yura, T. MRNA splicing-mediated c-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc. Natl. Acad. Sci. USA 2000, 97, 4660–4665. [Google Scholar] [CrossRef]
- Schroder, M.; Clark, R.; Kaufman, R.J. Ire1- and HAC1-independent transcriptional regulation in the unfolded protein response of yeast. Mol. Microbiol. 2003, 49, 591–606. [Google Scholar] [CrossRef]
- Sidrauski, C.; Chapman, R.; Walter, P. The unfolded protein response: An intracellular signalling pathway with many surprising features. Trends Cell Biol. 1998, 8, 245–249. [Google Scholar] [CrossRef]
- Anshu, A.; Mannan, M.A.; Chakraborty, A.; Chakrabarti, S.; Dey, M. A novel role for protein kinase kin2 in regulating HAC1 mRNA translocation, splicing, and translation. Mol. Cell Biol. 2015, 35, 199–210. [Google Scholar] [CrossRef]
- Ruegsegger, U.; Leber, J.H.; Walter, P. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell 2001, 107, 103–114. [Google Scholar] [CrossRef]
- Di Santo, R.; Aboulhouda, S.; Weinberg, D.E. The fail-safe mechanism of post-transcriptional silencing of unspliced HAC1 mRNA. eLife 2016, 5, e20069. [Google Scholar] [CrossRef] [PubMed]
- Kawahara, T.; Yanagi, H.; Yura, T.; Mori, K. Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/ern4p that activates the unfolded protein response. Mol. Biol. Cell 1997, 8, 1845–1862. [Google Scholar] [CrossRef] [PubMed]
- Sathe, L.; Bolinger, C.; Mannan, M.A.; Dever, T.E.; Dey, M. Evidence that base-pairing interaction between intron and mRNA leader sequences inhibits initiation of HAC1 mRNA translation in yeast. J. Biol. Chem. 2015, 290, 21821–21832. [Google Scholar] [CrossRef] [PubMed]
- Payne, T.; Hanfrey, C.; Bishop, A.L.; Michael, A.J.; Avery, S.V.; Archer, D.B. Transcript-specific translational regulation in the unfolded protein response of Saccharomyces cerevisiae. FEBS Lett. 2008, 582, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, K.M.; DeRisi, J.L.; Brown, P.O.; Sarnow, P. Global and specific translational regulation in the genomic response of Saccharomyces cerevisiae to a rapid transfer from a fermentable to a nonfermentable carbon source. Mol. Cell Biol. 2001, 21, 916–927. [Google Scholar] [CrossRef]
- Arava, Y.; Wang, Y.; Storey, J.D.; Liu, C.L.; Brown, P.O.; Herschlag, D. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2003, 100, 3889–3894. [Google Scholar] [CrossRef]
- Cox, J.S.; Shamu, C.E.; Walter, P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 1993, 73, 1197–1206. [Google Scholar] [CrossRef]
- Mori, K.; Ma, W.; Gething, M.J.; Sambrook, J. A transmembrane protein with a cdc2+/cdc28-related kinase activity is required for signaling from the ER to the nucleus. Cell 1993, 74, 743–756. [Google Scholar]
- Ishiwata-Kimata, Y.; Yamamoto, Y.H.; Takizawa, K.; Kohno, K.; Kimata, Y. F-actin and a type-ii myosin are required for efficient clustering of the ER stress sensor IRE1. Cell Struct. Funct. 2013, 38, 135–143. [Google Scholar] [CrossRef]
- Van Anken, E.; Pincus, D.; Coyle, S.; Aragon, T.; Osman, C.; Lari, F.; Gomez Puerta, S.; Korennykh, A.V.; Walter, P. Specificity in endoplasmic reticulum-stress signaling in yeast entails a step-wise engagement of HAC1 mRNA to clusters of the stress sensor IRE1. eLife 2014, 3, e05031. [Google Scholar] [CrossRef] [PubMed]
- Korennykh, A.V.; Egea, P.F.; Korostelev, A.A.; Finer-Moore, J.; Zhang, C.; Shokat, K.M.; Stroud, R.M.; Walter, P. The unfolded protein response signals through high-order assembly of IRE1. Nature 2009, 457, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Welihinda, A.A.; Tirasophon, W.; Green, S.R.; Kaufman, R.J. Protein serine/threonine phosphatase ptc2p negatively regulates the unfolded-protein response by dephosphorylating Ire1p kinase. Mol. Cell Biol. 1998, 18, 1967–1977. [Google Scholar] [CrossRef] [PubMed]
- Korennykh, A.V.; Korostelev, A.A.; Egea, P.F.; Finer-Moore, J.; Stroud, R.M.; Zhang, C.; Shokat, K.M.; Walter, P. Structural and functional basis for RNA cleavage by IRE1. BMC Biol. 2011, 9, 47. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, D.; Paira, S.; Das, B. Nuclear mRNA degradation tunes the gain of the unfolded protein response in Saccharomyces cerevisiae. Nucleic. Acids Res. 2018, 46, 1139–1156. [Google Scholar] [CrossRef]
- Pal, B.; Chan, N.C.; Helfenbaum, L.; Tan, K.; Tansey, W.P.; Gething, M.J. Scfcdc4-mediated degradation of the Hac1p transcription factor regulates the unfolded protein response in Saccharomyces cerevisiae. Mol. Biol. Cell 2007, 18, 426–440. [Google Scholar] [CrossRef]
- Mori, K. The unfolded protein response: The dawn of a new field. Proc. Jpn. Acad. Ser. B Physic. Biol. Sci. 2015, 91, 469–480. [Google Scholar] [CrossRef]
- Walter, P. Walking along the serendipitous path of discovery. Mol. Biol. Cell 2010, 21, 15–17. [Google Scholar] [CrossRef]
- Patil, C.K.; Li, H.; Walter, P. Gcn4p and novel upstream activating sequences regulate targets of the unfolded protein response. PLoS Biol. 2004, 2, E246. [Google Scholar] [CrossRef]
- Herzog, B.; Popova, B.; Jakobshagen, A.; Shahpasandzadeh, H.; Braus, G.H. Mutual cross talk between the regulators HAC1 of the unfolded protein response and gcn4 of the general amino acid control of Saccharomyces cerevisiae. Eukaryot. Cell. 2013, 12, 1142–1154. [Google Scholar] [CrossRef]
- Ogawa, N.; Mori, K. Autoregulation of the HAC1 gene is required for sustained activation of the yeast unfolded protein response. Genes Cells 2004, 9, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Polymenis, M. Dcr2 targets IRE1 and downregulates the unfolded protein response in Saccharomyces cerevisiae. EMBO Rep. 2006, 7, 1124–1127. [Google Scholar] [CrossRef] [PubMed]
- Kimata, Y.; Oikawa, D.; Shimizu, Y.; Ishiwata-Kimata, Y.; Kohno, K. A role for bip as an adjustor for the endoplasmic reticulum stress-sensing protein IRE1. J. Cell Biol. 2004, 167, 445–456. [Google Scholar] [CrossRef] [PubMed]
- Pincus, D.; Chevalier, M.W.; Aragon, T.; van Anken, E.; Vidal, S.E.; El-Samad, H.; Walter, P. Bip binding to the ER-stress sensor IRE1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol. 2010, 8, e1000415. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, D.; Kimata, Y.; Kohno, K. Self-association and bip dissociation are not sufficient for activation of the ER stress sensor IRE1. J. Cell Sci. 2007, 120, 1681–1688. [Google Scholar] [CrossRef] [PubMed]
- Pleiss, J.A.; Whitworth, G.B.; Bergkessel, M.; Guthrie, C. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol. Cell 2007, 27, 928–937. [Google Scholar] [CrossRef] [PubMed]
- Xia, X. Rna-seq approach for accurate characterization of splicing efficiency of yeast introns. Methods 2019. [Google Scholar] [CrossRef]
- Xia, X. Arsda: A new approach for storing, transmitting and analyzing transcriptomic data. G3 Genes Genom. Genet. 2017, 7, 3839–3848. [Google Scholar] [CrossRef]
- Kimata, Y.; Kohno, K. Endoplasmic reticulum stress-sensing mechanisms in yeast and mammalian cells. Curr. Opin. Cell. Biol. 2011, 23, 135–142. [Google Scholar] [CrossRef]
- Bertolotti, A.; Zhang, Y.; Hendershot, L.M.; Harding, H.P.; Ron, D. Dynamic interaction of bip and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2000, 2, 326–332. [Google Scholar] [CrossRef]
- Okamura, K.; Kimata, Y.; Higashio, H.; Tsuru, A.; Kohno, K. Dissociation of Kar2p/bip from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem. Biophys. Res. Commun. 2000, 279, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Credle, J.J.; Finer-Moore, J.S.; Papa, F.R.; Stroud, R.M.; Walter, P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2005, 102, 18773–18784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubio, C.; Pincus, D.; Korennykh, A.; Schuck, S.; El-Samad, H.; Walter, P. Homeostatic adaptation to endoplasmic reticulum stress depends on IRE1 kinase activity. J. Cell Biol. 2011, 193, 171–184. [Google Scholar] [CrossRef] [PubMed]
- Gardner, B.M.; Walter, P. Unfolded proteins are IRE1-activating ligands that directly induce the unfolded protein response. Science 2011, 333, 1891–1894. [Google Scholar] [CrossRef] [PubMed]
- Karagoz, G.E.; Acosta-Alvear, D.; Nguyen, H.T.; Lee, C.P.; Chu, F.; Walter, P. An unfolded protein-induced conformational switch activates mammalian IRE1. eLife 2017, 6, e30700. [Google Scholar] [CrossRef]
- Dong, L.; Krewson, E.A.; Yang, L.V. Acidosis activates endoplasmic reticulum stress pathways through gpr4 in human vascular endothelial cells. Int. J. Mol. Sci. 2017, 18, 278. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Nino, M.; Marquina, M.; Swinnen, S.; Rodriguez-Porrata, B.; Nevoigt, E.; Arino, J. The cytosolic ph of individual Saccharomyces cerevisiae cells is a key factor in acetic acid tolerance. Appl. Environ. Microbiol. 2015, 81, 7813–7821. [Google Scholar] [CrossRef] [PubMed]
- Valli, M.; Sauer, M.; Branduardi, P.; Borth, N.; Porro, D.; Mattanovich, D. Intracellular ph distribution in Saccharomyces cerevisiae cell populations, analyzed by flow cytometry. Appl. Environ. Microbiol. 2005, 71, 1515–1521. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [Green Version]
- Xia, X. Dambe7: New and improved tools for data analysis in molecular biology and evolution. Mol. Biol. Evol. 2018, 35, 1550–1552. [Google Scholar] [CrossRef]
- Mori, K.; Sant, A.; Kohno, K.; Normington, K.; Gething, M.J.; Sambrook, J.F. A 22 bp cis-acting element is necessary and sufficient for the induction of the yeast KAR2 (bip) gene by unfolded proteins. Embo J. 1992, 11, 2583–2593. [Google Scholar] [CrossRef] [PubMed]
- Van Dalfsen, K.M.; Hodapp, S.; Keskin, A.; Otto, G.M.; Berdan, C.A.; Higdon, A.; Cheunkarndee, T.; Nomura, D.K.; Jovanovic, M.; Brar, G.A. Global proteome remodeling during ER stress involves HAC1-driven expression of long undecoded transcript isoforms. Dev. Cell 2018, 46, 219–235. [Google Scholar] [CrossRef] [PubMed]
- Kohno, K.; Normington, K.; Sambrook, J.; Gething, M.J.; Mori, K. The promoter region of the yeast KAR2 (bip) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol. Cell Biol. 1993, 13, 877–890. [Google Scholar] [CrossRef] [PubMed]
- Chawla, A.; Chakrabarti, S.; Ghosh, G.; Niwa, M. Attenuation of yeast UPR is essential for survival and is mediated by IRE1 kinase. J. Cell Biol. 2011, 193, 41–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casagrande, R.; Stern, P.; Diehn, M.; Shamu, C.; Osario, M.; Zuniga, M.; Brown, P.O.; Ploegh, H. Degradation of proteins from the ER of s. Cerevisiae requires an intact unfolded protein response pathway. Mol. Cell 2000, 5, 729–735. [Google Scholar] [CrossRef]
- Arroyo, J.; Hutzler, J.; Bermejo, C.; Ragni, E.; Garcia-Cantalejo, J.; Botias, P.; Piberger, H.; Schott, A.; Sanz, A.B.; Strahl, S. Functional and genomic analyses of blocked protein o-mannosylation in baker’s yeast. Mol. Microbiol. 2011, 79, 1529–1546. [Google Scholar] [CrossRef] [PubMed]
- Bonilla, M.; Nastase, K.K.; Cunningham, K.W. Essential role of calcineurin in response to endoplasmic reticulum stress. Embo J. 2002, 21, 2343–2353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gardarin, A.; Chedin, S.; Lagniel, G.; Aude, J.C.; Godat, E.; Catty, P.; Labarre, J. Endoplasmic reticulum is a major target of cadmium toxicity in yeast. Mol. Microbiol. 2010, 76, 1034–1048. [Google Scholar] [CrossRef] [PubMed]
- Kawazoe, N.; Kimata, Y.; Izawa, S. Acetic acid causes endoplasmic reticulum stress and induces the unfolded protein response in Saccharomyces cerevisiae. Front. Microbiol. 2017, 8, 1192. [Google Scholar] [CrossRef]
- Chaillot, J.; Tebbji, F.; Remmal, A.; Boone, C.; Brown, G.W.; Bellaoui, M.; Sellam, A. The monoterpene carvacrol generates endoplasmic reticulum stress in the pathogenic fungus Candida albicans. Antimicrob. Agents Chemother 2015, 59, 4584–4592. [Google Scholar] [CrossRef]
- Burt, S.A.; van der Zee, R.; Koets, A.P.; de Graaff, A.M.; van Knapen, F.; Gaastra, W.; Haagsman, H.P.; Veldhuizen, E.J.A. Carvacrol Induces Heat Shock Protein 60 and Inhibits Synthesis of Flagellin in Escherichia coli O157:H7. Appl. Environ. Microbiol. 2007, 73, 4484. [Google Scholar] [CrossRef] [PubMed]
- Graf, A.; Gasser, B.; Dragosits, M.; Sauer, M.; Leparc, G.G.; Tuchler, T.; Kreil, D.P.; Mattanovich, D. Novel insights into the unfolded protein response using Pichia pastoris specific DNA microarrays. BMC Genom. 2008, 9, 390. [Google Scholar] [CrossRef]
- Iracane, E.; Donovan, P.D.; Ola, M.; Butler, G.; Holland, L.M. Identification of an exceptionally long intron in the HAC1 gene of Candida parapsilosis. mSphere 2018, 3, e00532-18. [Google Scholar] [CrossRef] [PubMed]
- Shechtman, C.F.; Henneberry, A.L.; Seimon, T.A.; Tinkelenberg, A.H.; Wilcox, L.J.; Lee, E.; Fazlollahi, M.; Munkacsi, A.B.; Bussemaker, H.J.; Tabas, I.; et al. Loss of subcellular lipid transport due to arv1 deficiency disrupts organelle homeostasis and activates the unfolded protein response. J. Biol. Chem. 2011, 286, 11951–11959. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Feldman, D.E.; Deng, C.; Brown, J.A.; De Giacomo, A.F.; Gaw, A.F.; Shi, G.; Le, Q.T.; Brown, J.M.; Koong, A.C. Identification of mitogen-activated protein kinase signaling pathways that confer resistance to endoplasmic reticulum stress in Saccharomyces cerevisiae. Mol. Cancer Res. 2005, 3, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Bertolotti, A.; Ron, D. Alterations in an IRE1-RNA complex in the mammalian unfolded protein response. J. Cell Sci. 2001, 114, 3207–3212. [Google Scholar] [PubMed]
- Armstrong, M.C.; Sestak, S.; Ali, A.A.; Sagini, H.A.M.; Brown, M.; Baty, K.; Treumann, A.; Schroder, M. Bypass of activation loop phosphorylation by aspartate 836 in activation of the endoribonuclease activity of IRE1. Mol. Cell Biol. 2017, 37. [Google Scholar] [CrossRef]
- Kimmig, P.; Diaz, M.; Zheng, J.; Williams, C.C.; Lang, A.; Aragon, T.; Li, H.; Walter, P. The unfolded protein response in fission yeast modulates stability of select mRNAs to maintain protein homeostasis. eLife 2012, 1, e00048. [Google Scholar] [CrossRef] [PubMed]
- Guydosh, N.R.; Kimmig, P.; Walter, P.; Green, R. Regulated IRE1-dependent mRNA decay requires no-go mRNA degradation to maintain endoplasmic reticulum homeostasis in S. pombe. eLife 2017, 6, e29216. [Google Scholar] [CrossRef] [PubMed]
- Hollien, J.; Lin, J.H.; Li, H.; Stevens, N.; Walter, P.; Weissman, J.S. Regulated IRE1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol. 2009, 186, 323–331. [Google Scholar] [CrossRef]
- Hollien, J.; Weissman, J.S. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science 2006, 313, 104–107. [Google Scholar] [CrossRef] [PubMed]
- Steffen, K.K.; McCormick, M.A.; Pham, K.M.; MacKay, V.L.; Delaney, J.R.; Murakami, C.J.; Kaeberlein, M.; Kennedy, B.K. Ribosome deficiency protects against ER stress in Saccharomyces cerevisiae. Genetics 2012, 191, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.A.; DiDone, L.; Krysan, D.J. Extracellular secretion of overexpressed glycosylphosphatidylinositol-linked cell wall protein utr2/crh2p as a novel protein quality control mechanism in Saccharomyces cerevisiae. Eukaryot. Cell 2010, 9, 1669–1679. [Google Scholar] [CrossRef] [PubMed]
- Travers, K.J.; Patil, C.K.; Wodicka, L.; Lockhart, D.J.; Weissman, J.S.; Walter, P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 2000, 101, 249–258. [Google Scholar] [CrossRef]
- Cakir, B. Bax induces activation of the unfolded protein response by inducing HAC1 mRNA splicing in Saccharomyces cerevisiae. Yeast 2012, 29, 395–406. [Google Scholar] [CrossRef]
- Han, D.; Lerner, A.G.; Vande Walle, L.; Upton, J.P.; Xu, W.; Hagen, A.; Backes, B.J.; Oakes, S.A.; Papa, F.R. Ire1alpha kinase activation modes control Alternate endoribonuclease outputs to determine divergent cell fates. Cell 2009, 138, 562–575. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Brandizzi, F. Ire1: Er stress sensor and cell fate executor. Trends Cell Biol. 2013, 23, 547–555. [Google Scholar] [CrossRef]
- Upton, J.P.; Wang, L.; Han, D.; Wang, E.S.; Huskey, N.E.; Lim, L.; Truitt, M.; McManus, M.T.; Ruggero, D.; Goga, A.; et al. Ire1alpha cleaves select microRNAs during ER stress to derepress translation of proapoptotic caspase-2. Science 2012, 338, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Niwa, M.; Patil, C.K.; DeRisi, J.; Walter, P. Genome-scale approaches for discovering novel nonconventional splicing substrates of the IRE1 nuclease. Genom. Biol. 2005, 6, R3. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, H.; Brandizzi, F.; Verchot, J.; Wang, A. The UPR branch IRE1-bZIP60 in plants plays an essential role in viral infection and is complementary to the only UPR pathway in yeast. PLoS Genet. 2015, 11, e1005164. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, C.; Wang, A. Divergence and conservation of the major UPR branch IRE1-bZIP signaling pathway across eukaryotes. Sci. Rep. 2016, 6, 27362. [Google Scholar] [CrossRef] [PubMed]
- Tirasophon, W.; Welihinda, A.A.; Kaufman, R.J. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998, 12, 1812–1824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, H.; Matsui, T.; Yamamoto, A.; Okada, T.; Mori, K. Xbp1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 2001, 107, 881–891. [Google Scholar] [CrossRef]
- Iwakoshi, N.N.; Lee, A.H.; Glimcher, L.H. The x-box binding protein-1 transcription factor is required for plasma cell differentiation and the unfolded protein response. Immunol. Rev. 2003, 194, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, S.; Wakasa, Y.; Takahashi, H.; Kawakatsu, T.; Takaiwa, F. Signal transduction by IRE1-mediated splicing of bzip50 and other stress sensors in the endoplasmic reticulum stress response of rice. Plant J. 2012, 69, 946–956. [Google Scholar] [CrossRef] [PubMed]
- Calfon, M.; Zeng, H.; Urano, F.; Till, J.H.; Hubbard, S.R.; Harding, H.P.; Clark, S.G.; Ron, D. Ire1 couples endoplasmic reticulum load to secretory capacity by processing the xbp-1 mRNA. Nature 2002, 415, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Foti, D.M.; Welihinda, A.; Kaufman, R.J.; Lee, A.S. Conservation and divergence of the yeast and mammalian unfolded protein response. Activation of specific mammalian endoplasmic reticulum stress element of the grp78/bip promoter by yeast HAC1. J. Biol. Chem. 1999, 274, 30402–30409. [Google Scholar] [CrossRef]
- Walter, P.; Ron, D. The unfolded protein response: From stress pathway to homeostatic regulation. Science 2011, 334, 1081–1086. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.J.; Strudwick, N.; Ali, A.A.; Paton, A.W.; Paton, J.C.; Schroder, M. Measuring signaling by the unfolded protein response. Methods Enzymol. 2011, 491, 261–292. [Google Scholar]
- Hernandez-Elvira, M.; Torres-Quiroz, F.; Escamilla-Ayala, A.; Dominguez-Martin, E.; Escalante, R.; Kawasaki, L.; Ongay-Larios, L.; Coria, R. The unfolded protein response pathway in the yeast kluyveromyces lactis. A comparative view among yeast species. Cells 2018, 7, 106. [Google Scholar] [CrossRef]
- Peschek, J.; Acosta-Alvear, D.; Mendez, A.S.; Walter, P. A conformational RNA zipper promotes intron ejection during non-conventional XBP1 mRNA splicing. EMBO Rep. 2015, 16, 1688–1698. [Google Scholar] [CrossRef] [PubMed]
- Hooks, K.B.; Griffiths-Jones, S. Conserved RNA structures in the non-canonical HAC1/XBP1 intron. RNA Biol. 2011, 8, 552–556. [Google Scholar] [CrossRef]
- Li, W.; Okreglak, V.; Peschek, J.; Kimmig, P.; Zubradt, M.; Weissman, J.S.; Walter, P. Engineering ER-stress dependent non-conventional mRNA splicing. eLife 2018, 7, e35388. [Google Scholar] [CrossRef] [PubMed]
- Iwawaki, T.; Tokuda, M. Function of yeast and amphioxus tRNA ligase in IRE1alpha-dependent XBP1 mRNA splicing. Biochem. Biophys. Res. Commun. 2011, 413, 527–531. [Google Scholar] [CrossRef]
- Jurkin, J.; Henkel, T.; Nielsen, A.F.; Minnich, M.; Popow, J.; Kaufmann, T.; Heindl, K.; Hoffmann, T.; Busslinger, M.; Martinez, J. The mammalian tRNA ligase complex mediates splicing of XBP1 mRNA and controls antibody secretion in plasma cells. Embo J. 2014, 33, 2922–2936. [Google Scholar] [CrossRef]
- Lu, Y.; Liang, F.X.; Wang, X. A synthetic biology approach identifies the mammalian UPR RNA ligase rtcb. Mol. Cell 2014, 55, 758–770. [Google Scholar] [CrossRef]
- Mori, T.; Ogasawara, C.; Inada, T.; Englert, M.; Beier, H.; Takezawa, M.; Endo, T.; Yoshihisa, T. Dual functions of yeast tRNA ligase in the unfolded protein response: Unconventional cytoplasmic splicing of HAC1 pre-mRNA is not sufficient to release translational attenuation. Mol. Biol. Cell 2010, 21, 3722–3734. [Google Scholar] [CrossRef]
- Oh, M.H.; Cheon, S.A.; Kang, H.A.; Kim, J.Y. Functional characterization of the unconventional splicing of yarrowia lipolytica HAC1 mRNA induced by unfolded protein response. Yeast 2010, 27, 443–452. [Google Scholar] [CrossRef]
- Mulder, H.J.; Nikolaev, I. Haca-dependent transcriptional switch releases haca mRNA from a translational block upon endoplasmic reticulum stress. Eukaryot. Cell 2009, 8, 665–675. [Google Scholar] [CrossRef] [PubMed]
- Saloheimo, M.; Valkonen, M.; Penttila, M. Activation mechanisms of the HAC1-mediated unfolded protein response in filamentous fungi. Mol. Microbiol. 2003, 47, 1149–1161. [Google Scholar] [CrossRef]
- Tirosh, B.; Iwakoshi, N.N.; Glimcher, L.H.; Ploegh, H.L. Rapid turnover of unspliced xbp-1 as a factor that modulates the unfolded protein response. J. Biol. Chem. 2006, 281, 5852–5860. [Google Scholar] [CrossRef] [PubMed]
- Bowring, C.E.; Llewellyn, D.H. Differences in HAC1 mRNA processing and translation between yeast and mammalian cells indicate divergence of the eukaryotic ER stress response. Biochem. Biophys. Res. Commun. 2001, 287, 789–800. [Google Scholar] [CrossRef] [PubMed]
- Niwa, M.; Sidrauski, C.; Kaufman, R.J.; Walter, P. A role for presenilin-1 in nuclear accumulation of IRE1 fragments and induction of the mammalian unfolded protein response. Cell 1999, 99, 691–702. [Google Scholar] [CrossRef]



| Gene | SystName | NEE | NEI5 | NEI3 |
|---|---|---|---|---|
| RPL36B | YPL249C-A | 3603 | 10 | 16 |
| RPL18A | YOL120C | 2789 | 10 | 11 |
| RPL39 | YJL189W | 7590 | 11 | 40 |
| RIM1 | YCR028C-A | 146 | 0 | 1 |
| RPL43A | YPR043W | 4083 | 14 | 15 |
| RPL40A | YIL148W | 3928 | 5 | 19 |
| RPL23B | YER117W | 3192 | 19 | 7 |
| RPL2B | YIL018W | 2974 | 4 | 14 |
| RPS21B | YJL136C | 4192 | 10 | 14 |
| RPL31B | YLR406C | 919 | 4 | 2 |
| RPL25 | YOL127W | 6536 | 13 | 22 |
| RPL31A | YDL075W | 3032 | 10 | 6 |
| RPL6A | YML073C | 3009 | 5 | 10 |
| RPS4B | YHR203C | 3411 | 13 | 6 |
| RPS21A | YKR057W | 2205 | 5 | 4 |
| RPS4A | YJR145C | 4206 | 8 | 10 |
| RPL33A | YPL143W | 4880 | 11 | 10 |
| SAC6 | YDR129C | 242 | 1 | 0 |
| RPS7A | YOR096W | 5093 | 4 | 13 |
| HAC1 | YFL031W | 32 | 844 | 704 |
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, X. Translation Control of HAC1 by Regulation of Splicing in Saccharomyces cerevisiae. Int. J. Mol. Sci. 2019, 20, 2860. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20122860
Xia X. Translation Control of HAC1 by Regulation of Splicing in Saccharomyces cerevisiae. International Journal of Molecular Sciences. 2019; 20(12):2860. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20122860
Chicago/Turabian StyleXia, Xuhua. 2019. "Translation Control of HAC1 by Regulation of Splicing in Saccharomyces cerevisiae" International Journal of Molecular Sciences 20, no. 12: 2860. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20122860