Unveiling the Interplay between the TLR4/MD2 Complex and HSP70 in the Human Cardiovascular System: A Computational Approach
Abstract
:1. Introduction
2. Results
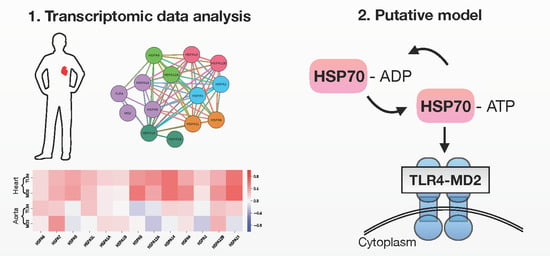
2.1. TLR4/MD2 Complex Transcriptomic Levels Correlate with HSP70 in Human Cardiovascular Tissues
2.2. Putative Interaction Model between the TLR4/MD2 Complex and HSP70
2.3. ATP Stimulates the Formation of Red Flurescence in PLA for HSP70 and TLR4
3. Discussion
4. Materials and Methods
4.1. Electrostatic Potential
4.2. Network Evidence of HSP70 and TLR4 Protein-Protein Interaction
4.3. RNA Sequence Data Analysis
4.4. Molecular Docking
4.5. Normal Mode Analysis
4.6. In Situ Proximity Ligation Assay
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Cardiovascular Diseases. 2017. Available online: https://www.who.int/cardiovascular_diseases (accessed on 21 October 2018).
- Mann, D. The emerging role of innate immunity in the heart and vascular system: For whom the cell tolls. Circ. Res. 2011, 108, 1133. [Google Scholar] [CrossRef] [PubMed]
- de Kleijn, D.; Pasterkamp, G. Toll-like receptors in cardiovascular diseases. Cardiovasc. Res. 2003, 60, 58–67. [Google Scholar] [CrossRef]
- Caso, J.R.; Pradillo, J.M.; Hurtado, O.; Lorenzo, P.; Moro, M.A.; Lizasoain, I. Toll-like receptor 4 is involved in brain damage and inflammation after experimental stroke. Circulation 2007, 115, 1599–1608. [Google Scholar] [CrossRef] [PubMed]
- Edfeldt, K.; Swedenborg, J.; Hansson, G.K.; Yan, Z.Q. Expression of toll-like receptors in human atherosclerotic lesions: A possible pathway for plaque activation. Circulation 2002, 105, 1158–1161. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, A.A.; Davis, D.; Nunes, K.P. Pattern recognition receptors as potential therapeutic targets in metabolic syndrome: From bench to bedside. Diabetes Metab. Syndr. Clin. Res. Rev. 2019, 13, 1117–1122. [Google Scholar] [CrossRef]
- Nunes, K.P.; de Oliveira, A.A.; Lima, V.V.; Webb, R.C. Toll-like receptor 4 and blood pressure: Lessons from animal studies. Front. Physiol. 2019, 10, 655. [Google Scholar] [CrossRef] [PubMed]
- Nunes, K.P.; Bomfim, G.F.; Toque, H.A.; Szasz, T.; Webb, R.C. Toll-like receptor 4 (TLR4) impairs nitric oxide contributing to Angiotensin II-induced cavernosal dysfunction. Life Sci. 2017, 191, 219–226. [Google Scholar] [CrossRef]
- Nunes, K.P.; de Oliveira, A.A.; Szasz, T.; Biancardi, V.C.; Webb, R.C. Blockade of Toll-Like Receptor 4 Attenuates Erectile Dysfunction in Diabetic Rats. J. Sex. Med. 2018, 15, 1235–1245. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Lv, J.; Jiang, S.; Ma, Z.; Wang, D.; Hu, W.; Deng, C.; Fan, C.; Di, S.; Sun, Y.; et al. The emerging role of Toll-like receptor 4 in myocardial inflammation. Cell Death Dis. 2017, 7, e2234. [Google Scholar] [CrossRef]
- Katare, P.B.; Bagul, P.K.; Dinda, A.K.; Banerjee, S.K. Toll-like receptor 4 inhibition improves oxidative stress and mitochondrial health in isoproterenol-induced cardiac hypertrophy in rats. Front. Immunol. 2017, 8, 719. [Google Scholar] [CrossRef] [PubMed]
- De Batista, P.R.; Palacios, R.; Martín, A.; Hernanz, R.; Médici, C.T.; Silva, M.A.; Rossi, E.M.; Aguado, A.; Vassallo, D.V.; Salaices, M.; et al. Toll-like receptor 4 upregulation by angiotensin II contributes to hypertension and vascular dysfunction through reactive oxygen species production. PLoS ONE 2014, 9, e104020. [Google Scholar] [CrossRef] [PubMed]
- Ehrentraut, H.; Ehrentraut, S.F.; Boehm, O.; El Aissati, S.; Foltz, F.; Goelz, L.; Goertz, D.; Kebir, S.; Weisheit, C.; Wolf, M.; et al. Tlr4 deficiency protects against cardiac pressure overload induced hyperinflammation. PLoS ONE 2015, 10, e0142921. [Google Scholar] [CrossRef] [PubMed]
- Nunes, K.P.; de Oliveira, A.A.; Mowry, F.E.; Biancardi, V.C. Targeting toll-like receptor 4 signalling pathways: Can therapeutics pay the toll for hypertension? Br. J. Pharmacol. 2018, 176, 1864–1879. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Wang, L.; Chen, S. Endogenous toll-like receptor ligands and their biological significance. J. Cell. Mol. Med. 2010, 14, 2592–2603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vabulas, R.M.; Ahmad-Nejad, P.; Ghose, S.; Kirschning, C.J.; Issels, R.D.; Wagner, H. HSP70 as endogenous stimulus of toll/interleukin-1 receptor signal pathway. J. Biol. Chem. 2002, 277, 15107–15112. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Wu, Y.; Huang, X.; Wang, W.; Ang, B.; Cao, X.; Wan, T. TLR4 is essential for HSP70-like protein 1 (HSP70L1) to activate dendritic cells and induce Th1 response. J. Biol. Chem. 2011, 286, 30393–30400. [Google Scholar] [CrossRef] [PubMed]
- Luong, M.; Zhang, Y.; Chamberlain, T.; Zhou, T.; Wright, J.F.; Dower, K.; Hall, J.P. Stimulation of TLR4 by recombinant HSP70 requires structural integrity of the HSP70 protein itself. J. Inflamm. 2012, 9, 11. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, A.; Webb, R.; Nunes, K. Toll-Like Receptor 4 and Heat-Shock Protein 70: Is it a New Target Pathway for Diabetic Vasculopathies? Curr. Drug Targets 2018, 20, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Kampinga, H.H.; Hageman, J.; Vos, M.J.; Kubota, H.; Tanguay, R.M.; Bruford, E.A.; Cheetham, M.E.; Chen, B.; Hightower, L.E. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones 2009, 14, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Fernández, M.R.; Valpuesta, J.M. Hsp70 chaperone: A master player in protein homeostasis. F1000Research 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Asea, A. Heat shock proteins and toll-like receptors. In Toll-Like Receptors (TLRs) and Innate Immunity; Springer: Berlin/Heidelberg, Germany, 2008; Volume 183, pp. 111–127. [Google Scholar]
- Asea, A.; Rehli, M.; Kabingu, E.; Boch, J.A.; Baré, O.; Auron, P.E.; Stevenson, M.A.; Calderwood, S.K. Novel signal transduction pathway utilized by extracellular HSP70: Role of TLR2 and TLR4. J. Biol. Chem. 2002, 277, 15028–15034. [Google Scholar] [CrossRef] [PubMed]
- Daugaard, M.; Rohde, M.; Jäättelä, M. The heat shock protein 70 family: Highly homologous proteins with overlapping and distinct functions. FEBS Lett. 2007, 581, 3702–3710. [Google Scholar] [CrossRef] [Green Version]
- Brocchieri, L.; De Macario, E.C.; Macario, A.J. hsp70 genes in the human genome: Conservation and differentiation patterns predict a wide array of overlapping and specialized functions. BMC Evol. Biol. 2008, 8, 19. [Google Scholar] [CrossRef] [PubMed]
- Radons, J. The human HSP70 family of chaperones: Where do we stand? Cell Stress Chaperones 2016, 21, 379–404. [Google Scholar] [CrossRef] [PubMed]
- Zuiderweg, E.R.; Bertelsen, E.B.; Rousaki, A.; Mayer, M.P.; Gestwicki, J.E.; Ahmad, A. Allostery in the Hsp70 chaperone proteins. In Molecular Chaperones; Springer: Berlin/Heidelberg, Germany, 2012; Volume 328, pp. 99–153. [Google Scholar]
- Stetz, G.; Verkhivker, G.M. Dancing through Life: Molecular Dynamics Simulations and Network-Centric Modeling of Allosteric Mechanisms in Hsp70 and Hsp110 Chaperone Proteins. PLoS ONE 2015, 10, e0143752. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.; Bukau, B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell. Mol. Life Sci. 2005, 62, 670. [Google Scholar] [CrossRef]
- Mayer, M.P. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem. Sci. 2013, 38, 507–514. [Google Scholar] [CrossRef]
- Zhuravleva, A.; Gierasch, L.M. Substrate-binding domain conformational dynamics mediate Hsp70 allostery. Proc. Natl. Acad. Sci. USA 2015, 112, E2865–E2873. [Google Scholar] [CrossRef]
- Grunwald, M.S.; Ligabue-Braun, R.; Souza, C.S.; Heimfarth, L.; Verli, H.; Gelain, D.P.; Moreira, J.C.F. Putative model for heat shock protein 70 complexation with receptor of advanced glycation end products through fluorescence proximity assays and normal mode analyses. Cell Stress Chaperones 2017, 22, 99–111. [Google Scholar] [CrossRef]
- Anwar, M.A.; Panneerselvam, S.; Shah, M.; Choi, S. Insights into the species-specific TLR4 signaling mechanism in response to Rhodobacter sphaeroides lipid A detection. Sci. Rep. 2015, 5, 7657. [Google Scholar] [CrossRef] [Green Version]
- Shimazu, R.; Akashi, S.; Ogata, H.; Nagai, Y.; Fukudome, K.; Miyake, K.; Kimoto, M. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J. Exp. Med. 1999, 189, 1777–1782. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Frostegård, J. Immunity, atherosclerosis and cardiovascular disease. BMC Med. 2013, 11, 117. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Ruiz, I. Immune system and cardiovascular disease. Nat. Rev. Cardiol. 2016, 13, 503. [Google Scholar] [CrossRef] [PubMed]
- Vallejo, J.G. Role of toll-like receptors in cardiovascular diseases. Clin. Sci. 2011, 121, 1–10. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. Signaling to NF-κB by Toll-like receptors. Trends Mol. Med. 2007, 13, 460–469. [Google Scholar] [CrossRef]
- Wang, L.; Li, D.; Yang, K.; Hu, Y.; Zeng, Q. Toll-like receptor-4 and mitogen-activated protein kinase signal system are involved in activation of dendritic cells in patients with acute coronary syndrome. Immunology 2008, 125, 122–130. [Google Scholar] [CrossRef]
- Park, B.S.; Song, D.H.; Kim, H.M.; Choi, B.S.; Lee, H.; Lee, J.O. The structural basis of lipopolysaccharide recognition by the TLR4–MD-2 complex. Nature 2009, 458, 1191–1195. [Google Scholar] [CrossRef]
- Bellini, S.; Barutta, F.; Mastrocola, R.; Imperatore, L.; Bruno, G.; Gruden, G. Heat shock proteins in vascular diabetic complications: Review and future perspective. Int. J. Mol. Sci. 2017, 18, 2709. [Google Scholar] [CrossRef]
- Rodriguez-Iturbe, B.; Lanaspa, M.A.; Johnson, R.J. The role of autoimmune reactivity induced by heat shock protein 70 in the pathogenesis of essential hypertension. Br. J. Pharmacol. 2018, 176, 1829–1838. [Google Scholar] [CrossRef]
- Krause, M.; Heck, T.G.; Bittencourt, A.; Scomazzon, S.P.; Newsholme, P.; Curi, R.; Homem de Bittencourt, P.I. The chaperone balance hypothesis: The importance of the extracellular to intracellular HSP70 ratio to inflammation-driven type 2 diabetes, the effect of exercise, and the implications for clinical management. Mediat. Inflamm. 2015, 2015, 249205. [Google Scholar] [CrossRef] [PubMed]
- Deguchi, A.; Tomita, T.; Ohto, U.; Takemura, K.; Kitao, A.; Akashi-Takamura, S.; Miyake, K.; Maru, Y. Eritoran inhibits S100A8-mediated TLR4/MD-2 activation and tumor growth by changing the immune microenvironment. Oncogene 2016, 35, 1445–1456. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; He, M.; VanPatten, S.; Al-Abed, Y. Mechanistic insights into high mobility group box-1 (HMGb1)-induced Toll-like receptor 4 (TLR4) dimer formation. J. Biomol. Struct. Dyn. 2018, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Garate, J.A.; Oostenbrink, C. Lipid A from lipopolysaccharide recognition: Structure, dynamics and cooperativity by molecular dynamics simulations. Proteins Struct. Funct. Bioinform. 2013, 81, 658–674. [Google Scholar] [CrossRef] [PubMed]
- Billod, J.M.; Lacetera, A.; Guzmán-Caldentey, J.; Martín-Santamaría, S. Computational approaches to toll-like receptor 4 modulation. Molecules 2016, 21, 994. [Google Scholar] [CrossRef] [PubMed]
- López-Blanco, J.R.; Aliaga, J.I.; Quintana-Ortí, E.S.; Chacón, P. iMODS: Internal coordinates normal mode analysis server. Nucleic Acids Res. 2014, 42, W271–W276. [Google Scholar] [CrossRef] [PubMed]
- Kmiecik, S.; Kouza, M.; Badaczewska-Dawid, A.; Kloczkowski, A.; Kolinski, A. Modeling of protein structural flexibility and large-scale dynamics: Coarse-grained simulations and elastic network models. Int. J. Mol. Sci. 2018, 19, 3496. [Google Scholar] [CrossRef] [PubMed]
- Sheu, S.Y.; Yang, D.Y.; Selzle, H.; Schlag, E. Energetics of hydrogen bonds in peptides. Proc. Natl. Acad. Sci. USA 2003, 100, 12683–12687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pace, C.N.; Fu, H.; Lee Fryar, K.; Landua, J.; Trevino, S.R.; Schell, D.; Thurlkill, R.L.; Imura, S.; Scholtz, J.M.; Gajiwala, K.; et al. Contribution of hydrogen bonds to protein stability. Protein Sci. 2014, 23, 652–661. [Google Scholar] [CrossRef]
- Bagchi, S.; Fredriksson, R.; Wallén-Mackenzie, Å. In situ proximity ligation assay (PLA). In ELISA. Methods in Molecular Biology; Hnasko, R., Ed.; Humana Press: New York, NY, USA, 2015; Volume 1318, pp. 149–159. [Google Scholar]
- Martine, P.; Chevriaux, A.; Derangère, V.; Apetoh, L.; Garrido, C.; Ghiringhelli, F.; Rébé, C. HSP70 is a negative regulator of NLRP3 inflammasome activation. Cell Death Dis. 2019, 10, 256. [Google Scholar] [CrossRef] [Green Version]
- Baker, N.A.; Sept, D.; Joseph, S.; Holst, M.J.; McCammon, J.A. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. USA 2001, 98, 10037–10041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeLano, W.L. The PyMOL Molecular Graphics System. 2002. Available online: http://www.pymol.org (accessed on 21 October 2018).
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, W665–W667. [Google Scholar] [CrossRef] [PubMed]
- Barabási, A.L.; Oltvai, Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004, 5, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Vidal, M.; Cusick, M.E.; Barabási, A.L. Interactome networks and human disease. Cell 2011, 144, 986–998. [Google Scholar] [CrossRef] [PubMed]
- Blondel, V.D.; Guillaume, J.L.; Lambiotte, R.; Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. Theory Exp. 2008, 2008, P10008. [Google Scholar] [CrossRef] [Green Version]
- Hartman, R.; Faustino, J.; Pinheiro, D.; Menezes, R. Assessing the Suitability of Network Community Detection to Available Meta-data Using Rank Stability. In Proceedings of the International Conference on Web Intelligence, Leipzig, Germany, 23–26 August 2017; ACM: New York, NY, USA, 2017; pp. 162–169. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. In Proceedings of the Third International Conference on Weblogs and Social Media, San Jose, CA, USA, 17–20 May 2009. [Google Scholar]
- Enache, O.M.; Lahr, D.L.; Natoli, T.E.; Litichevskiy, L.; Wadden, D.; Flynn, C.; Gould, J.; Asiedu, J.K.; Narayan, R.; Subramanian, A. The GCTx format and cmapPy, R, M packages: Resources for the optimized storage and integrated traversal of dense matrices of data and annotations. bioRxiv 2017, 227041. [Google Scholar] [CrossRef]
- Seabold, S.; Perktold, J. Statsmodels: Econometric and Statistical Modeling with Python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; pp. 57–61. [Google Scholar]
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Bertelsen, E.B.; Chang, L.; Gestwicki, J.E.; Zuiderweg, E.R. Solution conformation of wild-type E. Coli Hsp70 (DnaK) Chaperone Complexed ADP Substrate. Proc. Natl. Acad. Sci. USA 2009, 106, 8471–8476. [Google Scholar] [CrossRef]
- Kityk, R.; Kopp, J.; Sinning, I.; Mayer, M.P. Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol. Cell 2012, 48, 863–874. [Google Scholar] [CrossRef] [PubMed]
- Kozakov, D.; Hall, D.R.; Xia, B.; Porter, K.A.; Padhorny, D.; Yueh, C.; Beglov, D.; Vajda, S. The ClusPro web server for protein–protein docking. Nat. Protoc. 2017, 12, 255–278. [Google Scholar] [CrossRef] [PubMed]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef] [PubMed]
- Tange, O. GNU Parallel 2018; Ole Tange: Copenhagen, Denmark, 2018. [Google Scholar] [CrossRef]
- Grüning, B.; Dale, R.; Sjödin, A.; Rowe, J.; Chapman, B.A.; Tomkins-Tinch, C.H.; Valieris, R.; Koster, J. Bioconda: A sustainable and comprehensive software distribution for the life sciences. BioRxiv 2017, 207092. [Google Scholar] [CrossRef] [PubMed]
- Rees, R.; Ziessen, T.; Ralph, D.; Kell, P.; Moncada, S.; Cellek, S. Human and rabbit cavernosal smooth muscle cells express Rho-kinase. Int. J. Impot. Res. 2002, 14, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| Variable | Heart (n = 303) | Aorta (n = 299) | |
|---|---|---|---|
| Gender | Female | 101 | 107 |
| Male | 202 | 192 | |
| Age | 20–29 | 21 | 26 |
| 30–39 | 18 | 26 | |
| 40–49 | 50 | 51 | |
| 50–59 | 111 | 94 | |
| 60–69 | 96 | 97 | |
| 70–79 | 7 | 5 | |
| Hardy scale death | Fast | 84 | 76 |
| Intermediate | 13 | 15 | |
| Slow | 25 | 22 | |
| Ventilator case | 180 | 185 | |
| Model | TLR4/MD2 vs. HSP70-ADP | TLR4/MD2 vs. HSP70-ATP | ||
|---|---|---|---|---|
| Energy (kcal/mol) | Cluster Size | Energy (kcal/mol) | Cluster Size | |
| Model.000.00 | −10.29 | 42 | −22.60 | 47 |
| Model.000.01 | −20.29 | 36 | −10.43 | 37 |
| Model.000.02 | −9.94 | 32 | −20.23 | 33 |
| Model.000.03 | −7.86 | 25 | −14.10 | 31 |
| Model.000.04 | −10.13 | 23 | −17.91 | 27 |
| Model.000.05 | −8.72 | 21 | −13.85 | 24 |
| Model.000.06 | −13.56 | 21 | −26.70 | 24 |
| Model.000.07 | −11.00 | 20 | −30.45 | 23 |
| Model.000.08 | −14.79 | 20 | −15.02 | 22 |
| Model.000.09 | −20.92 | 18 | −21.39 | 20 |
| TLR4/MD2 vs. HSP70-ADP | TLR4/MD2 vs. HSP70-ATP | ||||
|---|---|---|---|---|---|
| Donor | Aceptor | Distance | Donor | Aceptor | Distance |
| ARG56.A:NH1 | GLU485.B:OE1 | 2.719 | ARG56.A:NH1 | GLU94.B:OE2 | 2.709 |
| ASN61.A:N | GLU439.B:OE2 | 3.107 | ARG56.A:NH2 | GLU94.B:OE2 | 2.772 |
| ARG261.A:NH1 | GLN507.B:OE1 | 2.718 | ARG261.A:NE | GLU89.B:OE2 | 2.868 |
| TYR285.A:OH | GLN505.B:OE1 | 2.730 | ARG261.A:NH2 | GLU89.B:OE1 | 3.000 |
| HIS431.B:ND1 | ASP289.A:O | 2.773 | ARG261.A:NH2 | GLU89.B:OE2 | 2.819 |
| LYS435.B:NZ | ASP255.A:OD1 | 2.544 | LYS268.A:NZ | GLN115.B:OE1 | 2.704 |
| HIS458.B:ND1 | ASP255.A:OD2 | 2.933 | TYR285.A:OH | GLU42.B:OE1 | 2.762 |
| ARG460.B:NH1 | ARG56.A:O | 2.609 | ARG547.A:NE | GLU79.B:OE1 | 3.129 |
| ARG460.B:NH2 | LYS55.A:O | 2.742 | ARG547.A:NH2 | GLU79.B:OE1 | 2.786 |
| ARG460.B:NH2 | GLN57.A:O | 2.794 | LYS577.A:NZ | SER76.B:OG | 2.742 |
| GLN484.B:NE2 | ARG56.A:O | 3.655 | LYS20.B:NZ | GLN248.A:OE1 | 2.688 |
| GLN484.B:NE2 | GLN57.A:OE1 | 2.711 | LYS20.B:NZ | ASP289.A:OD2 | 2.854 |
| TYR22.B:OH | THR291.A:OG1 | 2.647 | |||
| LYS47.B:NZ | GLN44.A:OE1 | 2.822 | |||
| ARG68.B:NH1 | TYR285.A:OH | 2.580 | |||
| ARG68.B:NH2 | HIS295.A:NE2 | 2.969 | |||
| ARG87.B:NH2 | ASN282.A:OD1 | 2.697 | |||
| SER100.B:OG | ASP540.A:OD1 | 2.860 | |||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Oliveira, A.A.; Faustino, J.; de Lima, M.E.; Menezes, R.; Nunes, K.P. Unveiling the Interplay between the TLR4/MD2 Complex and HSP70 in the Human Cardiovascular System: A Computational Approach. Int. J. Mol. Sci. 2019, 20, 3121. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20133121
de Oliveira AA, Faustino J, de Lima ME, Menezes R, Nunes KP. Unveiling the Interplay between the TLR4/MD2 Complex and HSP70 in the Human Cardiovascular System: A Computational Approach. International Journal of Molecular Sciences. 2019; 20(13):3121. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20133121
Chicago/Turabian Stylede Oliveira, Amanda Almeida, Josemar Faustino, Maria Elena de Lima, Ronaldo Menezes, and Kenia Pedrosa Nunes. 2019. "Unveiling the Interplay between the TLR4/MD2 Complex and HSP70 in the Human Cardiovascular System: A Computational Approach" International Journal of Molecular Sciences 20, no. 13: 3121. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20133121