Prospective Application of Activity-Based Proteomic Profiling in Vision Research-Potential Unique Insights into Ocular Protease Biology and Pathology
Abstract
:1. Introduction
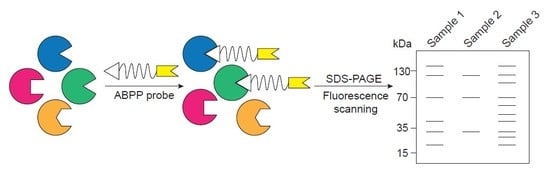
2. ABPP Experimental Design
3. Proteases in Eye Disease
3.1. Serine Proteases
3.2. Cysteine Proteases
3.3. Matrix Metalloproteinases
3.4. Other Proteases
4. Application of ABPP in Eye Research
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Wasinger, V.C.; Cordwell, S.J.; Cerpa-Poljak, A.; Yan, J.X.; Gooley, A.A.; Wilkins, M.R.; Duncan, M.W.; Harris, R.; Williams, K.L.; Humphery-Smith, I. Progress with gene-product mapping of the Mollicutes: Mycoplasma genitalium. Electrophoresis 1995, 16, 1090–1094. [Google Scholar] [CrossRef] [PubMed]
- Nagaraj, N.; Kulak, N.A.; Cox, J.; Neuhauser, N.; Mayr, K.; Hoerning, O.; Vorm, O.; Mann, M. System-wide perturbation analysis with nearly complete coverage of the yeast proteome by single-shot ultra HPLC runs on a bench top Orbitrap. Mol. Cell. Proteom. 2012, 11, M111.013722. [Google Scholar] [CrossRef] [PubMed]
- Picotti, P.; Clement-Ziza, M.; Lam, H.; Campbell, D.S.; Schmidt, A.; Deutsch, E.W.; Rost, H.; Sun, Z.; Rinner, O.; Reiter, L.; et al. A complete mass-spectrometric map of the yeast proteome applied to quantitative trait analysis. Nature 2013, 494, 266–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huttlin, E.L.; Jedrychowski, M.P.; Elias, J.E.; Goswami, T.; Rad, R.; Beausoleil, S.A.; Villen, J.; Haas, W.; Sowa, M.E.; Gygi, S.P. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 2010, 143, 1174–1189. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.-S.; Pinto, S.M.; Getnet, D.; Nirujogi, R.S.; Manda, S.S.; Chaerkady, R.; Madugundu, A.K.; Kelkar, D.S.; Isserlin, R.; Jain, S.; et al. A draft map of the human proteome. Nature 2014, 509, 575. [Google Scholar] [CrossRef] [PubMed]
- Lappalainen, T.; Sammeth, M.; Friedlander, M.R.; AC’t Hoen, P.A.; Monlong, J.; Rivas, M.A.; Gonzalez-Porta, M.; Kurbatova, N.; Griebel, T.; Ferreira, P.G.; et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 2013, 501, 506–511. [Google Scholar] [CrossRef] [PubMed]
- Pandey, A.; Mann, M. Proteomics to study genes and genomes. Nature 2000, 405, 837–846. [Google Scholar] [CrossRef]
- Cravatt, B.F.; Simon, G.M.; Yates, J.R., 3rd. The biological impact of mass-spectrometry-based proteomics. Nature 2007, 450, 991–1000. [Google Scholar] [CrossRef]
- Aebersold, R.; Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 2016, 537, 347–355. [Google Scholar] [CrossRef]
- Angel, T.E.; Aryal, U.K.; Hengel, S.M.; Baker, E.S.; Kelly, R.T.; Robinson, E.W.; Smith, R.D. Mass spectrometry-based proteomics: Existing capabilities and future directions. Chem. Soc. Rev. 2012, 41, 3912–3928. [Google Scholar] [CrossRef]
- Han, X.; Aslanian, A.; Yates, J.R., 3rd. Mass spectrometry for proteomics. Curr. Opin. Chem. Biol. 2008, 12, 483–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, J.D.; Schievella, A.R.; Nalefski, E.A.; Lin, L.L. Cytosolic phospholipase A2. J. Lipid Mediat. Cell Signal. 1995, 12, 83–117. [Google Scholar] [CrossRef]
- Mignatti, P.; Rifkin, D.B. Plasminogen activators and matrix metalloproteinases in angiogenesis. Enzym. Protein 1996, 49, 117–137. [Google Scholar] [CrossRef]
- Sanman, L.E.; Bogyo, M. Activity-based profiling of proteases. Annu. Rev. Biochem. 2014, 83, 249–273. [Google Scholar] [CrossRef] [PubMed]
- Nomura, D.K.; Dix, M.M.; Cravatt, B.F. Activity-based protein profiling for biochemical pathway discovery in cancer. Nat. Rev. Cancer 2010, 10, 630–638. [Google Scholar] [CrossRef] [Green Version]
- Jessani, N.; Liu, Y.; Humphrey, M.; Cravatt, B.F. Enzyme activity profiles of the secreted and membrane proteome that depict cancer cell invasiveness. Proc. Natl. Acad. Sci. USA 2002, 99, 10335–10340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wulfkuhle, J.D.; Paweletz, C.P.; Steeg, P.S.; Petricoin, E.F.; Liotta, L. Proteomic Approaches to the Diagnosis, Treatment, and Monitoring of Cancer. In New Trends in Cancer for the 21st Century: Proceedings of the International Symposium on Cancer: New Trends in Cancer for the 21st Century, held November 10–13, 2002, in Valencia, Spain; Llombart-Bosch, A., Felipo, V., Eds.; Springer: Boston, MA, USA, 2003; pp. 59–68. [Google Scholar] [CrossRef]
- Kim, S.I.; Voshol, H.; van Oostrum, J.; Hastings, T.G.; Cascio, M.; Glucksman, M.J. Neuroproteomics: Expression profiling of the brain’s proteomes in health and disease. Neurochem. Res. 2004, 29, 1317–1331. [Google Scholar] [CrossRef]
- Staub, I.; Sieber, S.A. β-Lactam Probes as Selective Chemical-Proteomic Tools for the Identification and Functional Characterization of Resistance Associated Enzymes in MRSA. J. Am. Chem. Soc. 2009, 131, 6271–6276. [Google Scholar] [CrossRef]
- Blais, D.R.; Brulotte, M.; Qian, Y.; Belanger, S.; Yao, S.Q.; Pezacki, J.P. Activity-based proteome profiling of hepatoma cells during hepatitis C virus replication using protease substrate probes. J. Proteome Res. 2010, 9, 912–923. [Google Scholar] [CrossRef]
- Semba, R.D.; Enghild, J.J. Proteomics and the eye. Proteom. Clin. Appl. 2014, 8, 127–129. [Google Scholar] [CrossRef]
- de Souza, G.A.; de Godoy, L.M.F.; Mann, M. Identification of 491 proteins in the tear fluid proteome reveals a large number of proteases and protease inhibitors. Genome Biol. 2006, 7, R72. [Google Scholar] [CrossRef] [PubMed]
- Dyrlund, T.F.; Poulsen, E.T.; Scavenius, C.; Nikolajsen, C.L.; Thogersen, I.B.; Vorum, H.; Enghild, J.J. Human cornea proteome: Identification and quantitation of the proteins of the three main layers including epithelium, stroma, and endothelium. J. Proteome Res. 2012, 11, 4231–4239. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, E.T.; Dyrlund, T.F.; Runager, K.; Scavenius, C.; Krogager, T.P.; Højrup, P.; Thøgersen, I.B.; Sanggaard, K.W.; Vorum, H.; Hjortdal, J.; et al. Proteomics of Fuchs’ Endothelial Corneal Dystrophy Support That the Extracellular Matrix of Descemet’s Membrane Is Disordered. J. Proteome Res. 2014, 13, 4659–4667. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Sawaguchi, S.; Twining, S.S.; Sugar, J.; Feder, R.S.; Yue, B.Y. Expression of degradative enzymes and protease inhibitors in corneas with keratoconus. Investig. Ophthalmol. Vis. Sci. 1998, 39, 1117–1124. [Google Scholar]
- Vaughan–Thomas, A.; Gilbert, S.J.; Duance, V.C. Elevated Levels of Proteolytic Enzymes in the Aging Human Vitreous. Investig. Ophthalmol. Vis. Sci. 2000, 41, 3299–3304. [Google Scholar]
- Gupta, V.; Mirzaei, M.; Gupta, V.B.; Chitranshi, N.; Dheer, Y.; Vander Wall, R.; Abbasi, M.; You, Y.; Chung, R.; Graham, S. Glaucoma is associated with plasmin proteolytic activation mediated through oxidative inactivation of neuroserpin. Sci. Rep. 2017, 7, 8412. [Google Scholar] [CrossRef] [PubMed]
- Wride, M.A.; Geatrell, J.; Guggenheim, J.A. Proteases in eye development and disease. Birth Defects Res. 2006, 78, 90–105. [Google Scholar] [CrossRef] [PubMed]
- Cooke Bailey, J.N.; Sobrin, L.; Pericak-Vance, M.A.; Haines, J.L.; Hammond, C.J.; Wiggs, J.L. Advances in the genomics of common eye diseases. Hum. Mol. Genet. 2013, 22, R59–R65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cravatt, B.F.; Wright, A.T.; Kozarich, J.W. Activity-based protein profiling: From enzyme chemistry to proteomic chemistry. Annu. Rev. Biochem. 2008, 77, 383–414. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Patricelli, M.P.; Cravatt, B.F. Activity-based protein profiling: The serine hydrolases. Proc. Natl. Acad. Sci. USA 1999, 96, 14694. [Google Scholar] [CrossRef] [PubMed]
- Saghatelian, A.; Jessani, N.; Joseph, A.; Humphrey, M.; Cravatt, B.F. Activity-based probes for the proteomic profiling of metalloproteases. Proc. Natl. Acad. Sci. USA 2004, 101, 10000–10005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, E.W.S.; Chattopadhaya, S.; Panicker, R.C.; Huang, X.; Yao, S.Q. Developing Photoactive Affinity Probes for Proteomic Profiling: Hydroxamate-based Probes for Metalloproteases. J. Am. Chem. Soc. 2004, 126, 14435–14446. [Google Scholar] [CrossRef] [PubMed]
- Weihofen, A.; Binns, K.; Lemberg, M.K.; Ashman, K.; Martoglio, B. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science 2002, 296, 2215–2218. [Google Scholar] [CrossRef] [PubMed]
- Fonovic, M.; Bogyo, M. Activity-based probes as a tool for functional proteomic analysis of proteases. Expert Rev. Proteom. 2008, 5, 721–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greenbaum, D.C.; Arnold, W.D.; Lu, F.; Hayrapetian, L.; Baruch, A.; Krumrine, J.; Toba, S.; Chehade, K.; Bromme, D.; Kuntz, I.D.; et al. Small molecule affinity fingerprinting. A tool for enzyme family subclassification, target identification, and inhibitor design. Chem. Biol. 2002, 9, 1085–1094. [Google Scholar] [CrossRef]
- Weerapana, E.; Speers, A.E.; Cravatt, B.F. Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)--a general method for mapping sites of probe modification in proteomes. Nat. Protoc. 2007, 2, 1414–1425. [Google Scholar] [CrossRef] [PubMed]
- Verhelst, S.H.; Fonovic, M.; Bogyo, M. A mild chemically cleavable linker system for functional proteomic applications. Angew. Chem. 2007, 46, 1284–1286. [Google Scholar] [CrossRef]
- López-Otín, C.; Bond, J.S. Proteases: Multifunctional Enzymes in Life and Disease. J. Biol. Chem. 2008, 283, 30433–30437. [Google Scholar] [CrossRef]
- Lutgens, S.P.M.; Cleutjens, K.B.J.M.; Daemen, M.J.A.P.; Heeneman, S. Cathepsin cysteine proteases in cardiovascular disease. Faseb J. 2007, 21, 3029–3041. [Google Scholar] [CrossRef]
- De Strooper, B. Proteases and Proteolysis in Alzheimer Disease: A Multifactorial View on the Disease Process. Physiol. Rev. 2010, 90, 465–494. [Google Scholar] [CrossRef]
- Pejler, G.; Rönnberg, E.; Waern, I.; Wernersson, S. Mast cell proteases: Multifaceted regulators of inflammatory disease. Blood 2010, 115, 4981. [Google Scholar] [CrossRef] [PubMed]
- Gold, B.; Merriam, J.E.; Zernant, J.; Hancox, L.S.; Taiber, A.J.; Gehrs, K.; Cramer, K.; Neel, J.; Bergeron, J.; Barile, G.R.; et al. Variation in factor B (BF) and complement component 2 (C2) genes is associated with age-related macular degeneration. Nat. Genet. 2006, 38, 458–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fagerness, J.A.; Maller, J.B.; Neale, B.M.; Reynolds, R.C.; Daly, M.J.; Seddon, J.M. Variation near complement factor I is associated with risk of advanced AMD. Eur. J. Hum. Genet. 2009, 17, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Geerlings, M.J.; de Jong, E.K.; den Hollander, A.I. The complement system in age-related macular degeneration: A review of rare genetic variants and implications for personalized treatment. Mol. Immunol. 2017, 84, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Montes, T.; Tortajada, A.; Morgan, B.P.; Rodriguez de Cordoba, S.; Harris, C.L. Functional basis of protection against age-related macular degeneration conferred by a common polymorphism in complement factor B. Proc. Natl. Acad. Sci. USA 2009, 106, 4366–4371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heurich, M.; Martinez-Barricarte, R.; Francis, N.J.; Roberts, D.L.; Rodriguez de Cordoba, S.; Morgan, B.P.; Harris, C.L. Common polymorphisms in C3, factor B, and factor H collaborate to determine systemic complement activity and disease risk. Proc. Natl. Acad. Sci. USA 2011, 108, 8761–8766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mantel, I.; Ambresin, A.; Moetteli, L.; Droz, I.; Roduit, R.; Munier, F.L.; Schorderet, D.F. Complement factor B polymorphism and the phenotype of early age-related macular degeneration. Ophthalmic Genet. 2014, 35, 12–17. [Google Scholar] [CrossRef]
- Hecker, L.A.; Edwards, A.O.; Ryu, E.; Tosakulwong, N.; Baratz, K.H.; Brown, W.L.; Charbel Issa, P.; Scholl, H.P.; Pollok-Kopp, B.; Schmid-Kubista, K.E.; et al. Genetic control of the alternative pathway of complement in humans and age-related macular degeneration. Hum. Mol. Genet. 2010, 19, 209–215. [Google Scholar] [CrossRef]
- Reynolds, R.; Hartnett, M.E.; Atkinson, J.P.; Giclas, P.C.; Rosner, B.; Seddon, J.M. Plasma complement components and activation fragments: Associations with age-related macular degeneration genotypes and phenotypes. Investig. Ophthalmol. Vis. Sci. 2009, 50, 5818–5827. [Google Scholar] [CrossRef]
- Loyet, K.M.; Deforge, L.E.; Katschke, K.J., Jr.; Diehl, L.; Graham, R.R.; Pao, L.; Sturgeon, L.; Lewin-Koh, S.C.; Hollyfield, J.G.; van Lookeren Campagne, M. Activation of the alternative complement pathway in vitreous is controlled by genetics in age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 2012, 53, 6628–6637. [Google Scholar] [CrossRef]
- Roversi, P.; Johnson, S.; Caesar, J.J.; McLean, F.; Leath, K.J.; Tsiftsoglou, S.A.; Morgan, B.P.; Harris, C.L.; Sim, R.B.; Lea, S.M. Structural basis for complement factor I control and its disease-associated sequence polymorphisms. Proc. Natl. Acad. Sci. USA 2011, 108, 12839–12844. [Google Scholar] [CrossRef] [Green Version]
- van de Ven, J.P.H.; Nilsson, S.C.; Tan, P.L.; Buitendijk, G.H.S.; Ristau, T.; Mohlin, F.C.; Nabuurs, S.B.; Schoenmaker-Koller, F.E.; Smailhodzic, D.; Campochiaro, P.A.; et al. A functional variant in the CFI gene confers a high risk of age-related macular degeneration. Nat. Genet. 2013, 45, 813. [Google Scholar] [CrossRef] [PubMed]
- Fritsche, L.G.; Chen, W.; Schu, M.; Yaspan, B.L.; Yu, Y.; Thorleifsson, G.; Zack, D.J.; Arakawa, S.; Cipriani, V.; Ripke, S.; et al. Seven new loci associated with age-related macular degeneration. Nat. Genet. 2013, 45, 433–439. [Google Scholar]
- Alexander, P.; Gibson, J.; Cree, A.J.; Ennis, S.; Lotery, A.J. Complement factor I and age-related macular degeneration. Mol. Vis. 2014, 20, 1253–1257. [Google Scholar] [PubMed]
- Seddon, J.M.; Yu, Y.; Miller, E.C.; Reynolds, R.; Tan, P.L.; Gowrisankar, S.; Goldstein, J.I.; Triebwasser, M.; Anderson, H.E.; Zerbib, J.; et al. Rare variants in CFI, C3 and C9 are associated with high risk of advanced age-related macular degeneration. Nat. Genet. 2013, 45, 1366. [Google Scholar] [CrossRef] [PubMed]
- Tan, P.L.; Garrett, M.E.; Willer, J.R.; Campochiaro, P.A.; Campochiaro, B.; Zack, D.J.; Ashley-Koch, A.E.; Katsanis, N. Systematic Functional Testing of Rare Variants: Contributions of CFI to Age-Related Macular DegenerationFunctional Rare Variants in CFI Contribute to AMD. Investig. Ophthalmol. Vis. Sci. 2017, 58, 1570–1576. [Google Scholar] [CrossRef]
- Kavanagh, D.; Yu, Y.; Schramm, E.C.; Triebwasser, M.; Wagner, E.K.; Raychaudhuri, S.; Daly, M.J.; Atkinson, J.P.; Seddon, J.M. Rare genetic variants in the CFI gene are associated with advanced age-related macular degeneration and commonly result in reduced serum factor I levels. Hum. Mol. Genet. 2015, 24, 3861–3870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, J.; Chen, Y.; Tong, Z.; Zhou, X.; Zhao, C.; Wang, K.; Hughes, G.; Kasuga, D.; Bedell, M.; Lee, C.; et al. Lack of association of CFD polymorphisms with advanced age-related macular degeneration. Mol. Vis. 2010, 16, 2273–2278. [Google Scholar]
- Stanton, C.M.; Yates, J.R.; den Hollander, A.I.; Seddon, J.M.; Swaroop, A.; Stambolian, D.; Fauser, S.; Hoyng, C.; Yu, Y.; Atsuhiro, K.; et al. Complement factor D in age-related macular degeneration. Investig. Ophthalmol. Vis. Sci. 2011, 52, 8828–8834. [Google Scholar] [CrossRef]
- Dewan, A.; Liu, M.; Hartman, S.; Zhang, S.S.; Liu, D.T.; Zhao, C.; Tam, P.O.; Chan, W.M.; Lam, D.S.; Snyder, M.; et al. HTRA1 promoter polymorphism in wet age-related macular degeneration. Science 2006, 314, 989–992. [Google Scholar] [CrossRef]
- Yang, Z.; Camp, N.J.; Sun, H.; Tong, Z.; Gibbs, D.; Cameron, D.J.; Chen, H.; Zhao, Y.; Pearson, E.; Li, X.; et al. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science 2006, 314, 992–993. [Google Scholar] [CrossRef] [PubMed]
- Deangelis, M.M.; Ji, F.; Adams, S.; Morrison, M.A.; Harring, A.J.; Sweeney, M.O.; Capone, A., Jr.; Miller, J.W.; Dryja, T.P.; Ott, J.; et al. Alleles in the HtrA serine peptidase 1 gene alter the risk of neovascular age-related macular degeneration. Ophthalmology 2008, 115, 1209–1215. [Google Scholar] [CrossRef] [PubMed]
- Clausen, T.; Kaiser, M.; Huber, R.; Ehrmann, M. HTRA proteases: Regulated proteolysis in protein quality control. Nat. Rev. Mol. Cell Biol. 2011, 12, 152. [Google Scholar] [CrossRef] [PubMed]
- Zurawa-Janicka, D.; Skorko-Glonek, J.; Lipinska, B. HtrA proteins as targets in therapy of cancer and other diseases. Expert Opin. Ther. Targets 2010, 14, 665–679. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.C.; Shen, D.; Zhou, M.; Ross, R.J.; Ding, X.; Zhang, K.; Green, W.R.; Tuo, J. Human HtrA1 in the archived eyes with age-related macular degeneration. Trans. Am. Ophthalmol. Soc. 2007, 105, 92–97. [Google Scholar] [PubMed]
- Lin, M.K.; Yang, J.; Hsu, C.W.; Gore, A.; Bassuk, A.G.; Brown, L.M.; Colligan, R.; Sengillo, J.D.; Mahajan, V.B.; Tsang, S.H. HTRA1, an age-related macular degeneration protease, processes extracellular matrix proteins EFEMP1 and TSP1. Aging Cell 2018, 17, e12710. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.; Kumar, S.; Zhang, N.; Tong, Z.; Yang, J.-H.; Watt, C.; Anderson, J.; Zhang, K.; Fillerup, H.; McCloskey, M.; et al. Increased expression of multifunctional serine protease, HTRA1, in retinal pigment epithelium induces polypoidal choroidal vasculopathy in mice. Proc. Natl. Acad. Sci. USA 2011, 108, 14578. [Google Scholar] [CrossRef]
- Oura, Y.; Nakamura, M.; Takigawa, T.; Fukushima, Y.; Wakabayashi, T.; Tsujikawa, M.; Nishida, K. High-Temperature Requirement A 1 Causes Photoreceptor Cell Death in Zebrafish Disease Models. Am. J. Pathol. 2018, 188, 2729–2744. [Google Scholar] [CrossRef] [Green Version]
- He, X.; Cheng, R.; Benyajati, S.; Ma, J.X. PEDF and its roles in physiological and pathological conditions: Implication in diabetic and hypoxia-induced angiogenic diseases. Clin. Sci. 2015, 128, 805–823. [Google Scholar] [CrossRef]
- Hulleman, J.D.; Genereux, J.C.; Nguyen, A. Mapping wild-type and R345W fibulin-3 intracellular interactomes. Exp. Eye Res. 2016, 153, 165–169. [Google Scholar] [CrossRef] [Green Version]
- Sathe, S.; Sakata, M.; Beaton, A.R.; Sack, R.A. Identification, origins and the diurnal role of the principal serine protease inhibitors in human tear fluid. Curr. Eye Res. 1998, 17, 348–362. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.Y.; Yang, X. Proteases for Cell Suicide: Functions and Regulation of Caspases. Microbiol. Mol. Biol. Rev. 2000, 64, 821. [Google Scholar] [CrossRef] [PubMed]
- Shalini, S.; Dorstyn, L.; Dawar, S.; Kumar, S. Old, new and emerging functions of caspases. Cell Death Differ. 2015, 22, 526–539. [Google Scholar] [CrossRef] [PubMed]
- McIlwain, D.R.; Berger, T.; Mak, T.W. Caspase functions in cell death and disease. Cold Spring Harb. Perspect. Biol. 2013, 5, a008656. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.N.; Berry, M.; Logan, A.; Blanch, R.J.; Ahmed, Z. Caspases in retinal ganglion cell death and axon regeneration. Cell Death Discov. 2017, 3, 17032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, C.N.; Thompson, A.M.; McCance, E.; Berry, M.; Logan, A.; Blanch, R.J.; Ahmed, Z. Caspase-2 Mediates Site-Specific Retinal Ganglion Cell Death After Blunt Ocular InjuryCaspase-2 in Blunt Ocular Injury. Investig. Ophthalmol. Vis. Sci. 2018, 59, 4453–4462. [Google Scholar] [CrossRef] [PubMed]
- Zacks, D.N.; Hӓnninen, V.; Pantcheva, M.; Ezra, E.; Grosskreutz, C.; Miller, J.W. Caspase Activation in an Experimental Model of Retinal Detachment. Investig. Ophthalmol. Vis. Sci. 2003, 44, 1262–1267. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, S.; Liu, Y.; Clark, A.F.; Pang, I.-H. Caspase-7: A critical mediator of optic nerve injury-induced retinal ganglion cell death. Mol. Neurodegener. 2015, 10, 40. [Google Scholar] [CrossRef]
- Mohr, S.; Xi, X.; Tang, J.; Kern, T.S. Caspase Activation in Retinas of Diabetic and Galactosemic Mice and Diabetic Patients. Diabetes 2002, 51, 1172. [Google Scholar] [CrossRef]
- Zhou, X.; Li, F.; Kong, L.; Tomita, H.; Li, C.; Cao, W. Involvement of inflammation, degradation, and apoptosis in a mouse model of glaucoma. J. Biol. Chem. 2005, 280, 31240–31248. [Google Scholar] [CrossRef]
- McKinnon, S.J.; Lehman, D.M.; Kerrigan-Baumrind, L.A.; Merges, C.A.; Pease, M.E.; Kerrigan, D.F.; Ransom, N.L.; Tahzib, N.G.; Reitsamer, H.A.; Levkovitch-Verbin, H.; et al. Caspase Activation and Amyloid Precursor Protein Cleavage in Rat Ocular Hypertension. Investig. Ophthalmol. Vis. Sci. 2002, 43, 1077–1087. [Google Scholar]
- Ahmed, Z.; Kalinski, H.; Berry, M.; Almasieh, M.; Ashush, H.; Slager, N.; Brafman, A.; Spivak, I.; Prasad, N.; Mett, I.; et al. Ocular neuroprotection by siRNA targeting caspase-2. Cell Death Dis. 2011, 2, e173. [Google Scholar] [CrossRef] [PubMed]
- Monnier, P.P.; Onofrio, P.M.; Magharious, M.; Hollander, A.C.; Tassew, N.; Szydlowska, K.; Tymianski, M.; Koeberle, P.D. Involvement of Caspase-6 and Caspase-8 in Neuronal Apoptosis and the Regenerative Failure of Injured Retinal Ganglion Cells. J. Neurosci. 2011, 31, 10494. [Google Scholar] [CrossRef] [PubMed]
- Beasley, S.; El-Sherbiny, M.; Megyerdi, S.; El-Shafey, S.; Choksi, K.; Kaddour-Djebbar, I.; Sheibani, N.; Hsu, S.; Al-Shabrawey, M. Caspase-14 Expression Impairs Retinal Pigment Epithelium Barrier Function: Potential Role in Diabetic Macular Edema. Biomed Res. Int. 2014, 2014, 11. [Google Scholar] [CrossRef] [PubMed]
- Kerur, N.; Fukuda, S.; Banerjee, D.; Kim, Y.; Fu, D.; Apicella, I.; Varshney, A.; Yasuma, R.; Fowler, B.J.; Baghdasaryan, E.; et al. cGAS drives noncanonical-inflammasome activation in age-related macular degeneration. Nat. Med. 2018, 24, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Ildefonso, C.J.; Jaime, H.; Biswal, M.R.; Boye, S.E.; Li, Q.; Hauswirth, W.W.; Lewin, A.S. Gene Therapy With the Caspase Activation and Recruitment Domain Reduces the Ocular Inflammatory Response. Mol. Ther. 2015, 23, 875–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Momeni, H.R. Role of calpain in apoptosis. Cell J. 2011, 13, 65–72. [Google Scholar]
- Potz, B.A.; Abid, M.R.; Sellke, F.W. Role of Calpain in Pathogenesis of Human Disease Processes. J. Nat. Sci. 2016, 2, e218. [Google Scholar]
- Perrin, B.J.; Huttenlocher, A. Calpain. Int. J. Biochem. Cell Biol. 2002, 34, 722–725. [Google Scholar] [CrossRef]
- Huang, W.; Fileta, J.; Rawe, I.; Qu, J.; Grosskreutz, C.L. Calpain Activation in Experimental Glaucoma. Investig. Ophthalmol. Vis. Sci. 2010, 51, 3049–3054. [Google Scholar] [CrossRef]
- Nakajima, E.; Hammond, K.B.; Rosales, J.L.; Shearer, T.R.; Azuma, M. Calpain, not caspase, is the causative protease for hypoxic damage in cultured monkey retinal cells. Investig. Ophthalmol. Vis. Sci. 2011, 52, 7059–7067. [Google Scholar] [CrossRef] [PubMed]
- Hoang, M.V.; Nagy, J.A.; Fox, J.E.; Senger, D.R. Moderation of calpain activity promotes neovascular integration and lumen formation during VEGF-induced pathological angiogenesis. PLoS ONE 2010, 5, e13612. [Google Scholar] [CrossRef] [PubMed]
- Hoang, M.V.; Smith, L.E.; Senger, D.R. Calpain inhibitors reduce retinal hypoxia in ischemic retinopathy by improving neovascular architecture and functional perfusion. Biochim. Et Biophys. Acta 2011, 1812, 549–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, Y.J.; Kim, M.S.; Chung, S.K. Calpain and Caspase-12 Expression in Lens Epithelial Cells of Diabetic Cataracts. Am. J. Ophthalmol. 2016, 167, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Im, E.; Kazlauskas, A. The role of cathepsins in ocular physiology and pathology. Exp. Eye Res. 2007, 84, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Turk, B.; Turk, D.; Turk, V. Lysosomal cysteine proteases: More than scavengers. Biochim. Et Biophys. Acta 2000, 1477, 98–111. [Google Scholar] [CrossRef]
- Brookes, N.H.; Loh, I.P.; Clover, G.M.; Poole, C.A.; Sherwin, T. Involvement of corneal nerves in the progression of keratoconus. Exp. Eye Res. 2003, 77, 515–524. [Google Scholar] [CrossRef]
- Sherwin, T.; Brookes, N.H.; Loh, I.P.; Poole, C.A.; Clover, G.M. Cellular incursion into Bowman’s membrane in the peripheral cone of the keratoconic cornea. Exp. Eye Res. 2002, 74, 473–482. [Google Scholar] [CrossRef]
- Kenney, M.C.; Chwa, M.; Atilano, S.R.; Tran, A.; Carballo, M.; Saghizadeh, M.; Vasiliou, V.; Adachi, W.; Brown, D.J. Increased levels of catalase and cathepsin V/L2 but decreased TIMP-1 in keratoconus corneas: Evidence that oxidative stress plays a role in this disorder. Investig. Ophthalmol. Vis. Sci. 2005, 46, 823–832. [Google Scholar] [CrossRef]
- Alizadeh, P.; Smit-McBride, Z.; Oltjen, S.L.; Hjelmeland, L.M. Regulation of cysteine cathepsin expression by oxidative stress in the retinal pigment epithelium/choroid of the mouse. Exp. Eye Res. 2006, 83, 679–687. [Google Scholar] [CrossRef] [Green Version]
- Bernstein, H.G.; Reichenbach, A.; Wiederanders, B. Cathepsin E immunoreactivity in human ocular tissues: Influence of aging and pathological states. Neurosci. Lett. 1998, 240, 135–138. [Google Scholar] [CrossRef]
- el-Hifnawi, E. Localization of cathepsin D in rat ocular tissues. An immunohistochemical study. Ann. Anat. 1995, 177, 11–17. [Google Scholar] [CrossRef]
- Lai, C.M.; Shen, W.Y.; Constable, I.; Rakoczy, P.E. The use of adenovirus-mediated gene transfer to develop a rat model for photoreceptor degeneration. Investig. Ophthalmol. Vis. Sci. 2000, 41, 580–584. [Google Scholar]
- Zurdel, J.; Finckh, U.; Menzer, G.; Nitsch, R.M.; Richard, G. CST3 genotype associated with exudative age related macular degeneration. Br. J. Ophthalmol. 2002, 86, 214–219. [Google Scholar] [CrossRef]
- Nguyen, A.; Hulleman, J.D. Evidence of Alternative Cystatin C Signal Sequence Cleavage Which Is Influenced by the A25T Polymorphism. PLoS ONE 2016, 11, e0147684. [Google Scholar] [CrossRef] [PubMed]
- Löffek, S.; Schilling, O.; Franzke, C.W. Biological role of matrix metalloproteinases: A critical balance. Eur. Respir. J. 2011, 38, 191. [Google Scholar] [CrossRef] [PubMed]
- Nagase, H.; Visse, R.; Murphy, G. Structure and function of matrix metalloproteinases and TIMPs. Cardiovasc. Res. 2006, 69, 562–573. [Google Scholar] [CrossRef] [Green Version]
- Brew, K.; Nagase, H. The tissue inhibitors of metalloproteinases (TIMPs): An ancient family with structural and functional diversity. Biochim. Et Biophys. Acta 2010, 1803, 55–71. [Google Scholar] [CrossRef] [Green Version]
- Singh, M.; Tyagi, S.C. Metalloproteinases as mediators of inflammation and the eyes: Molecular genetic underpinnings governing ocular pathophysiology. Int. J. Ophthalmol. 2017, 10, 1308–1318. [Google Scholar]
- Nita, M.; Strzalka-Mrozik, B.; Grzybowski, A.; Mazurek, U.; Romaniuk, W. Age-related macular degeneration and changes in the extracellular matrix. Med Sci. Monit. 2014, 20, 1003–1016. [Google Scholar] [Green Version]
- Hussain, A.A.; Lee, Y.; Zhang, J.-J.; Marshall, J. Disturbed Matrix Metalloproteinase Activity of Bruch’s Membrane in Age-Related Macular Degeneration. Investig. Ophthalmol. Vis. Sci. 2011, 52, 4459–4466. [Google Scholar] [CrossRef] [PubMed]
- Chau, K.Y.; Sivaprasad, S.; Patel, N.; Donaldson, T.A.; Luthert, P.J.; Chong, N.V. Plasma levels of matrix metalloproteinase-2 and -9 (MMP-2 and MMP-9) in age-related macular degeneration. Eye 2007, 22, 855. [Google Scholar] [CrossRef]
- Fiotti, N.; Pedio, M.; Battaglia Parodi, M.; Altamura, N.; Uxa, L.; Guarnieri, G.; Giansante, C.; Ravalico, G. MMP-9 microsatellite polymorphism and susceptibility to exudative form of age-related macular degeneration. Genet. Med. 2005, 7, 272–277. [Google Scholar] [CrossRef] [Green Version]
- Seitzman, R.L.; Mahajan, V.B.; Mangione, C.; Cauley, J.A.; Ensrud, K.E.; Stone, K.L.; Cummings, S.R.; Hochberg, M.C.; Hillier, T.A.; Sinsheimer, J.S.; et al. Estrogen receptor alpha and matrix metalloproteinase 2 polymorphisms and age-related maculopathy in older women. Am. J. Epidemiol. 2008, 167, 1217–1225. [Google Scholar] [CrossRef] [PubMed]
- Kowluru, R.A.; Zhong, Q.; Santos, J.M. Matrix metalloproteinases in diabetic retinopathy: Potential role of MMP-9. Expert Opin. Investig. Drugs 2012, 21, 797–805. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; McLamore, A.; Song, W.; McGuire, P.G. Retinal Neovascularization Is Suppressed With a Matrix Metalloproteinase Inhibitor. JAMA Ophthalmol. 1999, 117, 498–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giebel, S.J.; Menicucci, G.; McGuire, P.G.; Das, A. Matrix metalloproteinases in early diabetic retinopathy and their role in alteration of the blood-retinal barrier. Lab. Investig. 2005, 85, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, G.; Kowluru, R.A. Matrix metalloproteinase-2 in the development of diabetic retinopathy and mitochondrial dysfunction. Lab. Investig. 2010, 90, 1365. [Google Scholar] [CrossRef] [PubMed]
- De Groef, L.; Van Hove, I.; Dekeyster, E.; Stalmans, I.; Moons, L. MMPs in the trabecular meshwork: Promising targets for future glaucoma therapies? Investig. Ophthalmol. Vis. Sci. 2013, 54, 7756–7763. [Google Scholar] [CrossRef] [PubMed]
- Schlӧtzer-Schrehardt, U.; Lommatzsch, J.R.; Küchle, M.; Konstas, A.G.P.; Naumann, G.O.H. Matrix Metalloproteinases and Their Inhibitors in Aqueous Humor of Patients with Pseudoexfoliation Syndrome/Glaucoma and Primary Open-Angle Glaucoma. Investig. Ophthalmol. Vis. Sci. 2003, 44, 1117–1125. [Google Scholar] [CrossRef] [Green Version]
- Sahay, P.; Rao, A.; Padhy, D.; Sarangi, S.; Das, G.; Reddy, M.M.; Modak, R. Functional Activity of Matrix Metalloproteinases 2 and 9 in Tears of Patients with GlaucomaMMPs in Tear Film in Glaucoma. Investig. Ophthalmol. Vis. Sci. 2017, 58, BIO106–BIO113. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.Y.; Wu, Y.; Zhang, Y.; Liu, C.Y.; Deng, C.Y.; Peng, L.; Zhou, L. Associations between matrix metalloproteinase gene polymorphisms and glaucoma susceptibility: A meta-analysis. BMC Ophthalmol. 2017, 17, 48. [Google Scholar] [CrossRef] [PubMed]
- Liutkeviciene, R.; Lesauskaite, V.; Sinkunaite-Marsalkiene, G.; Zaliuniene, D.; Zaliaduonyte-Peksiene, D.; Mizariene, V.; Gustiene, O.; Jasinskas, V.; Jariene, G.; Tamosiunas, A. The Role of Matrix Metalloproteinases Polymorphisms in Age-Related Macular Degeneration. Ophthalmic Genet. 2015, 36, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Xia, W. gamma-Secretase and its modulators: Twenty years and beyond. Neurosci. Lett. 2019, 701, 162–169. [Google Scholar] [CrossRef]
- Maia, M.A.; Sousa, E. BACE-1 and gamma-Secretase as Therapeutic Targets for Alzheimer’s Disease. Pharm 2019, 12, 41. [Google Scholar]
- Qian, Q.; Mitter, S.K.; Pay, S.L.; Qi, X.; Rickman, C.B.; Grant, M.B.; Boulton, M.E. A Non-Canonical Role for beta-Secretase in the Retina. Adv. Exp. Med. Biol. 2016, 854, 333–339. [Google Scholar]
- Fielden, M.R.; Werner, J.; Jamison, J.A.; Coppi, A.; Hickman, D.; Dunn, R.T., 2nd; Trueblood, E.; Zhou, L.; Afshari, C.A.; Lightfoot-Dunn, R. Retinal Toxicity Induced by a Novel beta-secretase Inhibitor in the Sprague-Dawley Rat. Toxicol. Pathol. 2015, 43, 581–592. [Google Scholar] [CrossRef]
- Blasiak, J.; Pawlowska, E.; Szczepanska, J.; Kaarniranta, K. Interplay between Autophagy and the Ubiquitin-Proteasome System and Its Role in the Pathogenesis of Age-Related Macular Degeneration. Int. J. Mol. Sci. 2019, 20, 210. [Google Scholar] [CrossRef]
- Liu, K.; Lyu, L.; Chin, D.; Gao, J.; Sun, X.; Shang, F.; Caceres, A.; Chang, M.-L.; Rowan, S.; Peng, J.; et al. Altered ubiquitin causes perturbed calcium homeostasis, hyperactivation of calpain, dysregulated differentiation, and cataract. Proc. Natl. Acad. Sci. USA 2015, 112, 1071. [Google Scholar] [CrossRef]
- Shang, F.; Taylor, A. Role of the ubiquitin-proteasome in protein quality control and signaling: Implication in the pathogenesis of eye diseases. Prog. Mol. Biol. Transl. Sci. 2012, 109, 347–396. [Google Scholar]
- Zhao, Y.; Wilmarth, P.A.; Cheng, C.; Limi, S.; Fowler, V.M.; Zheng, D.; David, L.L.; Cvekl, A. Proteome-transcriptome analysis and proteome remodeling in mouse lens epithelium and fibers. Exp. Eye Res. 2018, 179, 32–46. [Google Scholar] [CrossRef] [PubMed]
- Azkargorta, M.; Soria, J.; Acera, A.; Iloro, I.; Elortza, F. Human tear proteomics and peptidomics in ophthalmology: Toward the translation of proteomic biomarkers into clinical practice. J. Proteom. 2017, 150, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Dor, M.; Eperon, S.; Lalive, P.H.; Guex-Crosier, Y.; Hamedani, M.; Salvisberg, C.; Turck, N. Investigation of the global protein content from healthy human tears. Exp. Eye Res. 2018, 179, 64–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eastlake, K.; Heywood, W.E.; Banerjee, P.; Bliss, E.; Mills, K.; Khaw, P.T.; Charteris, D.; Limb, G.A. Comparative proteomic analysis of normal and gliotic PVR retina and contribution of Muller glia to this profile. Exp. Eye Res. 2018, 177, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Ji, Y.W.; Hwang, H.S.; Oh, J.W.; Kim, H.C.; Lee, H.K.; Kim, K.P. Proteomic analysis of human lacrimal and tear fluid in dry eye disease. Sci. Rep. 2017, 7, 13363. [Google Scholar] [CrossRef]
- Murthy, K.R.; Goel, R.; Subbannayya, Y.; Jacob, H.K.C.; Murthy, P.R.; Manda, S.S.; Patil, A.H.; Sharma, R.; Sahasrabuddhe, N.A.; Parashar, A.; et al. Proteomic analysis of human vitreous humor. Clin. Proteom. 2014, 11, 29. [Google Scholar] [CrossRef] [PubMed]
- Schori, C.; Trachsel, C.; Grossmann, J.; Zygoula, I.; Barthelmes, D.; Grimm, C. The Proteomic Landscape in the Vitreous of Patients With Age-Related and Diabetic Retinal Disease. Investig. Ophthalmol. Vis. Sci. 2018, 59, Amd31–Amd40. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhu, X.; Lu, Y. The Proteome of Cataract Markers: Focus on Crystallins. Adv. Clin. Chem. 2018, 86, 179–210. [Google Scholar] [PubMed]
- Picciani, R.; Junk, A.K.; Bhattacharya, S.K. Technical Brief: A novel strategy for enrichment of trabecular meshwork protease proteome. Mol. Vis. 2008, 14, 871–877. [Google Scholar] [PubMed]
- Basavarajappa, H.D.; Lee, B.; Lee, H.; Sulaiman, R.S.; An, H.; Magaña, C.; Shadmand, M.; Vayl, A.; Rajashekhar, G.; Kim, E.-Y.; et al. Synthesis and Biological Evaluation of Novel Homoisoflavonoids for Retinal Neovascularization. J. Med. Chem. 2015, 58, 5015–5027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sulaiman, R.S.; Merrigan, S.; Quigley, J.; Qi, X.; Lee, B.; Boulton, M.E.; Kennedy, B.; Seo, S.Y.; Corson, T.W. A novel small molecule ameliorates ocular neovascularisation and synergises with anti-VEGF therapy. Sci. Rep. 2016, 6, 25509. [Google Scholar] [CrossRef] [PubMed]
- Sulaiman, R.S.; Park, B.; Sheik Pran Babu, S.P.; Si, Y.; Kharwadkar, R.; Mitter, S.K.; Lee, B.; Sun, W.; Qi, X.; Boulton, M.E.; et al. Chemical Proteomics Reveals Soluble Epoxide Hydrolase as a Therapeutic Target for Ocular Neovascularization. ACS Chem. Biol. 2018, 13, 45–52. [Google Scholar] [CrossRef] [PubMed]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, H.; Hulleman, J.D. Prospective Application of Activity-Based Proteomic Profiling in Vision Research-Potential Unique Insights into Ocular Protease Biology and Pathology. Int. J. Mol. Sci. 2019, 20, 3855. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20163855
Peng H, Hulleman JD. Prospective Application of Activity-Based Proteomic Profiling in Vision Research-Potential Unique Insights into Ocular Protease Biology and Pathology. International Journal of Molecular Sciences. 2019; 20(16):3855. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20163855
Chicago/Turabian StylePeng, Hui, and John D. Hulleman. 2019. "Prospective Application of Activity-Based Proteomic Profiling in Vision Research-Potential Unique Insights into Ocular Protease Biology and Pathology" International Journal of Molecular Sciences 20, no. 16: 3855. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20163855