E-cadherin Downregulation and microRNAs in Sporadic Intestinal-Type Gastric Cancer
Abstract
:1. Introduction
2. Results
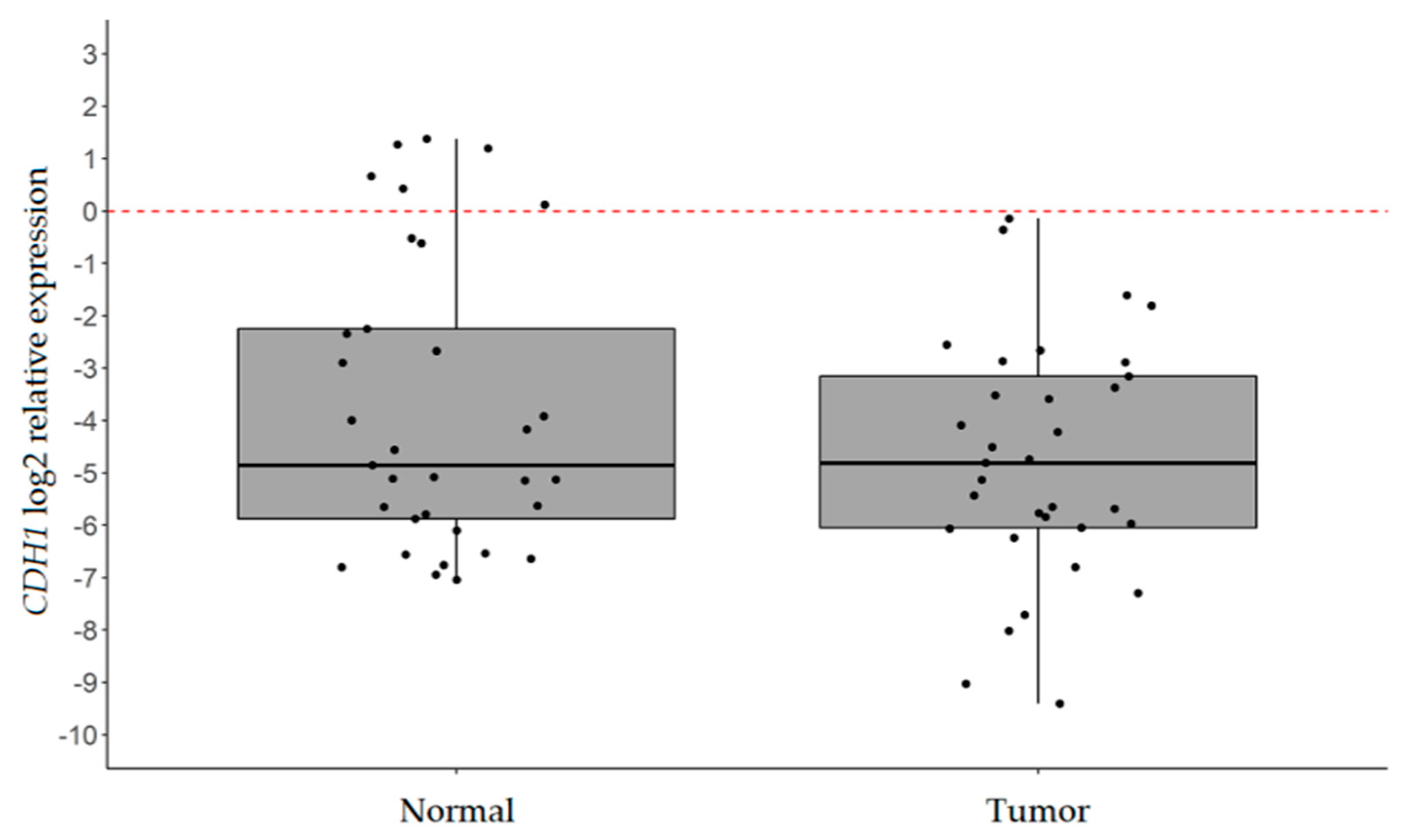
2.1. CDH1 is Significantly Downregulated in IGC
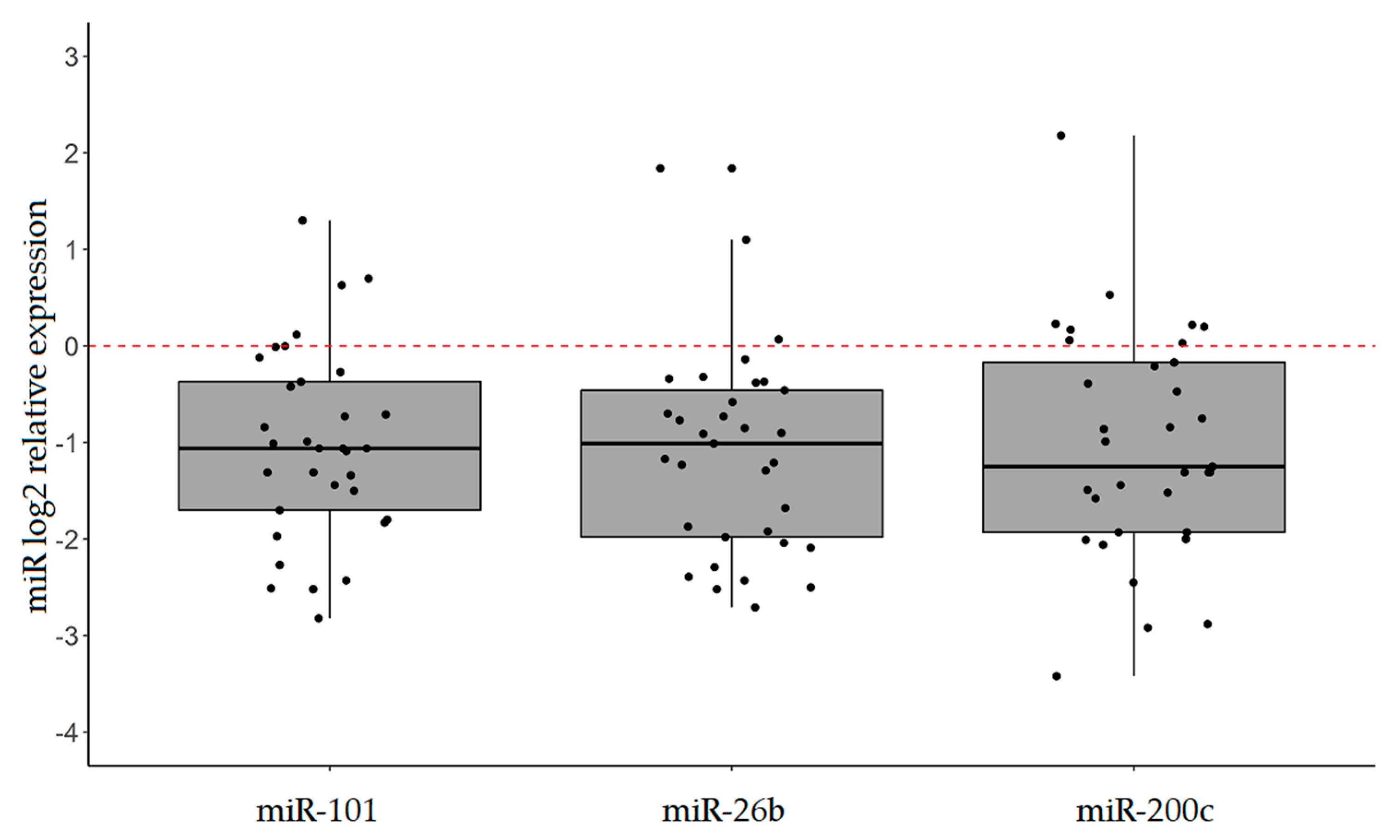
2.2. Explorative Analysis Suggests that the Expression of miR-101, miR-26b, and miR-200c is Altered in IGC
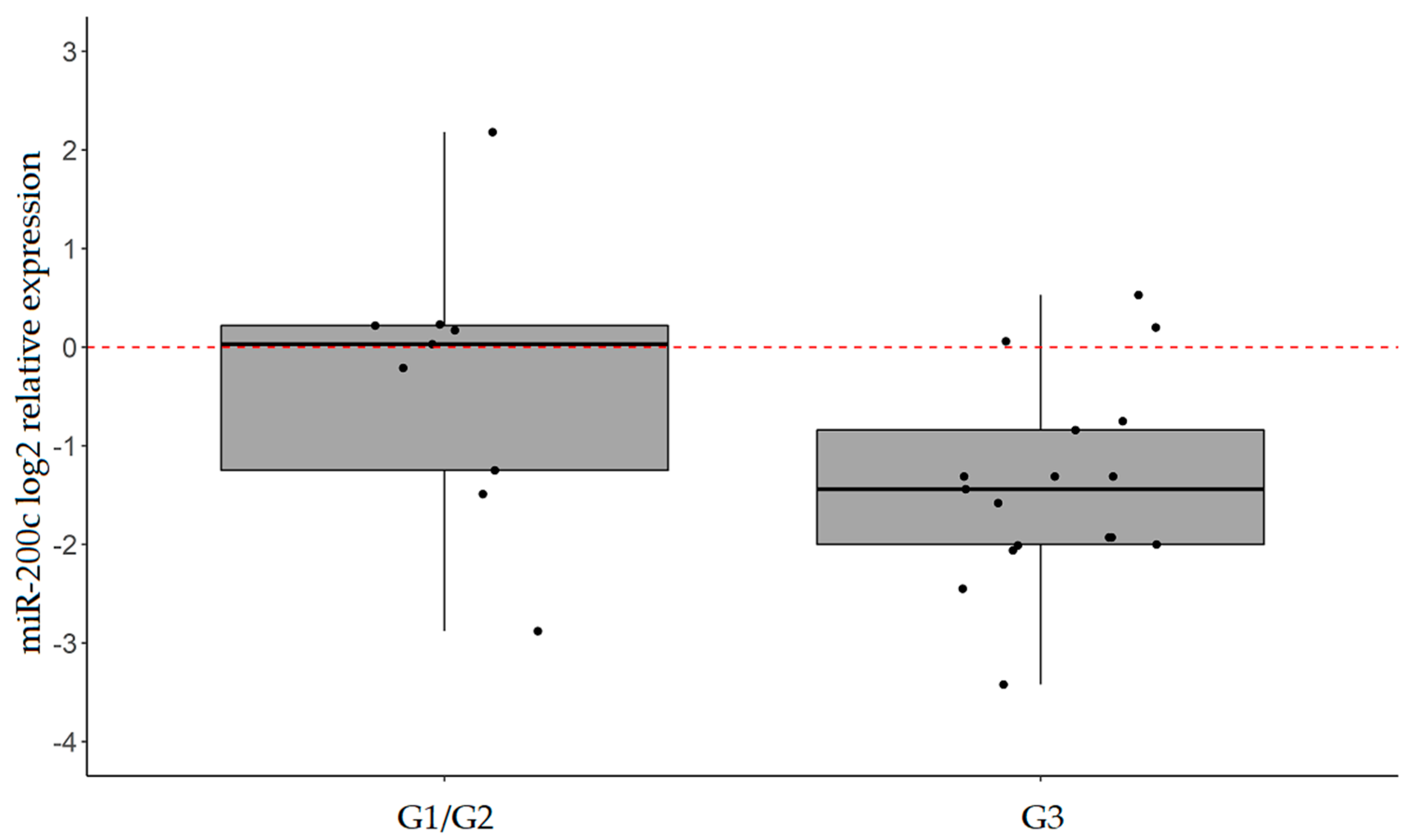
2.3. miR-101, miR-26b, and miR-200c are Confirmed to be Significantly Downregulated in IGC
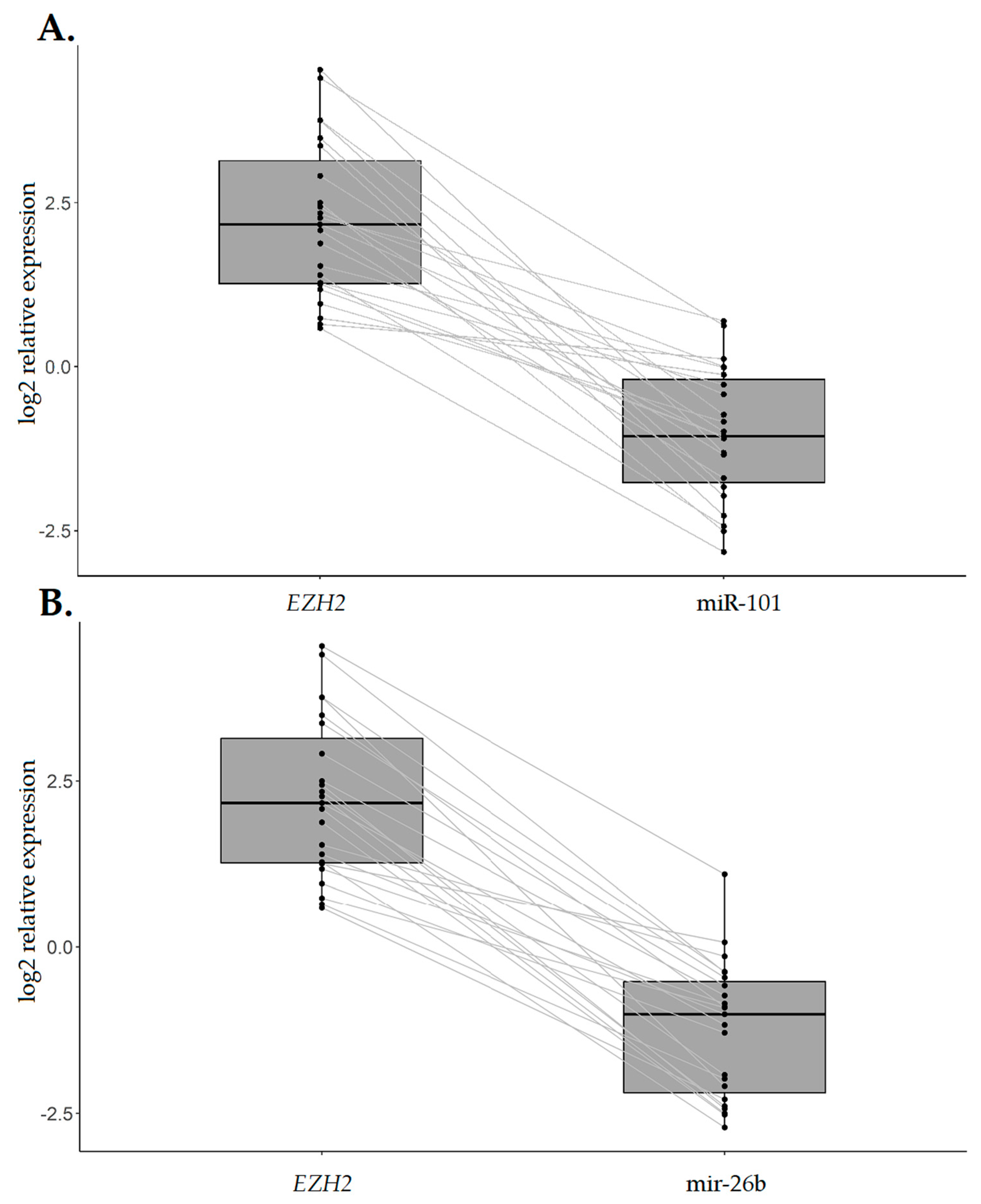
2.4. EZH2 Expression Levels are Increased in IGC Specimens Compared with the Normal Counterpart and Inversely Associated with CDH1 Expression
3. Discussion
4. Materials and Methods
4.1. Samples
4.2. RNA Extraction
4.3. Gene Expression Analyses
4.4. miR Analyses
4.5. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lauren, P. The two histological main types of gastric carcinoma: Diffuse and so-called intestinal-type carcinoma: An attempt at a histo-clinical classification. Acta Pathol. Microbiol. Scand. 1965, 64, 31–49. [Google Scholar] [CrossRef] [PubMed]
- Vauhkonen, M.; Vauhkonen, H.; Sipponen, P. Pathology and molecular biology of gastric cancer. Best Pract. Res. Clin. Gastroenterol. 2006, 20, 651–674. [Google Scholar] [CrossRef] [PubMed]
- Takeichi, M. Morphogenetic roles of classic cadherins. Curr. Opin. Cell Biol. 1995, 7, 619–627. [Google Scholar] [CrossRef]
- Gumbiner, B.M. Cell adhesion: The molecular basis of tissue architecture and morphogenesis. Cell 1996, 84, 345–357. [Google Scholar] [CrossRef]
- Takeichi, M. Cadherins in cancer: Implications for invasion and metastasis. Curr. Opin. Cell Biol. 1993, 5, 806–811. [Google Scholar] [CrossRef]
- Cano, A.; Pérez-Moreno, M.A.; Rodrigo, I.; Locascio, A.; Blanco, M.J.; del Barrio, M.G.; Portillo, F.; Nieto, M.A. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000, 2, 76–83. [Google Scholar] [CrossRef]
- Onder, T.T.; Gupta, P.B.; Mani, S.A.; Yang, J.; Lander, E.S.; Weinberg, R.A. Loss of E-Cadherin Promotes Metastasis via Multiple Downstream Transcriptional Pathways. Cancer Res. 2008, 68, 3645–3654. [Google Scholar] [CrossRef]
- Birchmeier, W. E-cadherin as a tumor (invasion) suppressor gene. Bioessays 1995, 17, 97–99. [Google Scholar] [CrossRef]
- Shimada, S.; Mimata, A.; Sekine, M.; Mogushi, K.; Akiyama, Y.; Fukamachi, H.; Jonkers, J.; Tanaka, H.; Eishi, Y.; Yuasa, Y. Synergistic tumour suppressor activity of E-cadherin and p53 in a conditional mouse model for metastatic diffuse-type gastric cancer. Gut 2012, 61, 344–353. [Google Scholar] [CrossRef]
- Carneiro, F.; Oliveira, C.; Suriano, G.; Seruca, R. Molecular pathology of familial gastric cancer, with an emphasis on hereditary diffuse gastric cancer. J. Clin. Pathol. 2008, 61, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Corso, G.; Carvalho, J.; Marrelli, D.; Vindigni, C.; Carvalho, B.; Seruca, R.; Roviello, F.; Oliveira, C. Somatic Mutations and Deletions of the E-Cadherin Gene Predict Poor Survival of Patients With Gastric Cancer. J. Clin. Oncol. 2013, 31, 868–875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Jansson, M.D.; Lund, A.H. MicroRNA and cancer. Mol. Oncol. 2012, 6, 590–610. [Google Scholar] [CrossRef] [PubMed]
- Juzėnas, S.; Saltenienė, V.; Kupcinskas, J.; Link, A.; Kiudelis, G.; Jonaitis, L.; Jarmalaite, S.; Kupcinskas, L.; Malfertheiner, P.; Skieceviciene, J. Analysis of Deregulated microRNAs and Their Target Genes in Gastric Cancer. PLoS ONE 2015, 10, e0132327. [Google Scholar] [CrossRef] [PubMed]
- Li, F.Q.; Xu, B.; Wu, Y.J.; Yang, Z.L.; Qian, J.J. Differential microRNA expression in signet-ring cell carcinoma compared with tubular adenocarcinoma of human gastric cancer. Genet. Mol. Res. 2015, 14, 739–747. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Chen, X.; Alattar, M.; Wei, J.; Liu, H. MicroRNAs in tumorigenesis, metastasis, diagnosis and prognosis of gastric cancer. Cancer Gene Ther. 2015, 22, 291–301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, J.; Min, B.H.; Jang, J.; Kang, S.Y.; Bae, H.; Jang, S.S.; Kim, J.I.; Kim, K.M. MicroRNA Expression Profiles in Gastric Carcinogenesis. Sci. Rep. 2018, 8, 14393. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ma, L. MicroRNA control of epithelial-mesenchymal transition and metastasis. Cancer Metastasis Rev. 2012, 31, 653–662. [Google Scholar] [CrossRef]
- De Craene, B.; Berx, G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer 2013, 13, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Bure, I.V.; Nemtsova, M.V.; Zaletaev, D.V. Roles of E-cadherin and Noncoding RNAs in the Epithelial–mesenchymal Transition and Progression in Gastric Cancer. Int. J. Mol. Sci. 2019, 20, 2870. [Google Scholar] [CrossRef] [PubMed]
- Díaz-López, A.; Moreno-Bueno, G.; Cano, A. Role of microRNA in epithelial to mesenchymal transition and metastasis and clinical perspectives. Cancer Manag. Res. 2014, 6, 205–216. [Google Scholar] [PubMed]
- Abba, M.; Patil, N.; Leupold, J.; Allgayer, H. MicroRNA Regulation of Epithelial to Mesenchymal Transition. J. Clin. Med. 2016, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chu, K.M. E-Cadherin and Gastric Cancer: Cause, Consequence, and Applications. Biomed Res. Int. 2014, 2014, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gan, W.J.; Wang, J.R.; Zhu, X.L.; He, X.S.; Guo, P.D.; Zhang, S.; Li, X.M.; Li, J.M.; Wu, H. RARγ-induced E-cadherin downregulation promotes hepatocellular carcinoma invasion and metastasis. J. Exp. Clin. Cancer Res. 2016, 35, 164. [Google Scholar] [CrossRef] [PubMed]
- Torabizadeh, Z.; Nosrati, A.; Sajadi Saravi, S.N.; Yazdani Charati, J.; Janbabai, G. Evaluation of E-cadherin Expression in Gastric Cancer and Its Correlation with Clinicopathologic Parameters. Int. J. Hematol. Oncol. Stem Cell Res. 2017, 11, 158–164. [Google Scholar] [PubMed]
- Ogasawara, N.; Kudo, T.; Sato, M.; Kawasaki, Y.; Yonezawa, S.; Takahashi, S.; Miyagi, Y.; Natori, Y.; Sugiyama, A. Reduction of Membrane Protein CRIM1 Decreases E-Cadherin and Increases Claudin-1 and MMPs, Enhancing the Migration and Invasion of Renal Carcinoma Cells. Biol. Pharm. Bull. 2018, 41, 604–611. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.A.; Inamura, K.; Yamauchi, M.; Nishihara, R.; Mima, K.; Sukawa, Y.; Li, T.; Yasunari, M.; Morikawa, T.; Fitzgerald, K.C.; et al. Loss of CDH1 (E-cadherin) expression is associated with infiltrative tumour growth and lymph node metastasis. Br. J. Cancer 2016, 114, 199–206. [Google Scholar] [CrossRef]
- Karayiannakis, A.J.; Syrigos, K.N.; Chatzigianni, E.; Papanikolaou, S.; Karatzas, G. E-cadherin expression as a differentiation marker in gastric cancer. Hepatogastroenterology 1998, 45, 2437–2442. [Google Scholar]
- Zhang, J.; Liu, X.; Yu, G.; Liu, L.; Wang, J.; Chen, X.; Bian, Y.; Ji, Y.; Zhou, X.; Chen, Y.; et al. UBE2C is a potential biomarker of intestinal-type gastric cancer with chromosomal instability. Front. Pharmacol. 2018, 9, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Croce, C.M. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012, 4, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Noto, J.M.; Peek, R.M. The Role of microRNAs in Helicobacter pylori Pathogenesis and Gastric Carcinogenesis. Front. Cell. Infect. Microbiol. 2012, 1, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, H.; Ma, Y.; Yang, M.; Zhang, C.; Huang, H.; Xia, Y.; Lu, L.; Jin, W.; Cui, D. Persisting and increasing neutrophil infiltration associates with gastric carcinogenesis and e-cadherin downregulation. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Ye, M.; Zhou, J.; Wang, Z.; Zhu, X. Targeting E-cadherin expression with small molecules for digestive cancer treatment. Am. J. Transl. Res. 2019, 11, 3932–3944. [Google Scholar] [PubMed]
- Abou Khouzam, R.; Molinari, C.; Salvi, S.; Marabelli, M.; Molinaro, V.; Orioli, D.; Saragoni, L.; Morgagni, P.; Calistri, D.; Ranzani, G.N. Digital PCR identifies changes in CDH1 (E-cadherin) transcription pattern in intestinal-type gastric cancer. Oncotarget 2017, 8, 18811–18820. [Google Scholar] [CrossRef]
- Varambally, S.; Cao, Q.; Mani, R.S.; Shankar, S.; Wang, X.; Ateeq, B.; Laxman, B.; Cao, X.; Jing, X.; Ramnarayanan, K.; et al. Genomic Loss of microRNA-101 Leads to Overexpression of Histone Methyltransferase EZH2 in Cancer. Science 2008, 322, 1695–1699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, F.; Cogdell, D.; Hu, L.; Yang, D.; Sood, A.K.; Xue, F.; Zhang, W. miR-101 suppresses the epithelial-to-mesenchymal transition by targeting ZEB1 and ZEB2 in ovarian carcinoma. Oncol. Rep. 2014, 31, 2021–2028. [Google Scholar] [CrossRef]
- Carvalho, J.; Van Grieken, N.C.; Pereira, P.M.; Sousa, S.; Tijssen, M.; Buffart, T.E.; Diosdado, B.; Grabsch, H.; Santos, M.A.S.; Meijer, G.; et al. Lack of microRNA-101 causes E-cadherin functional deregulation through EZH2 up-regulation in intestinal gastric cancer. J. Pathol. 2012, 228, 31–44. [Google Scholar] [CrossRef]
- Dong, X.; Liu, Y. Expression and significance of miR-24 and miR-101 in patients with advanced gastric cancer. Oncol. Lett. 2018, 16, 5769–5774. [Google Scholar] [CrossRef]
- Imamura, T.; Komatsu, S.; Ichikawa, D.; Miyamae, M.; Okajima, W.; Ohashi, T.; Kiuchi, J.; Nishibeppu, K.; Kosuga, T.; Konishi, H.; et al. Low plasma levels of miR-101 are associated with tumor progression in gastric cancer. Oncotarget 2017, 8, 106538. [Google Scholar] [CrossRef] [PubMed]
- Matsushima, K.; Isomoto, H.; Inoue, N.; Nakayama, T.; Hayashi, T.; Nakayama, M.; Nakao, K.; Hirayama, T.; Kohno, S. MicroRNA signatures in Helicobacter pylori-infected gastric mucosa. Int. J. Cancer 2011, 128, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Xia, Y.; Li, L.; Zhang, G. MiR-101 inhibits cell growth and tumorigenesis of Helicobacter pylori related gastric cancer by repression of SOCS2. Cancer Biol. Ther. 2015, 16, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Tsai, M.M.; Huang, H.W.; Wang, C.S.; Lee, K.F.; Tsai, C.Y.; Lu, P.H.; Chi, H.C.; Lin, Y.H.; Kuo, L.M.; Lin, K.H. MicroRNA-26b inhibits tumor metastasis by targeting the KPNA2/c-jun pathway in human gastric cancer. Oncotarget 2016, 7, 39511–39526. [Google Scholar] [CrossRef] [PubMed]
- Deng, M.; Zhang, R.; He, Z.; Qiu, Q.; Lu, X.; Yin, J.; Liu, H.; Jia, X.; He, Z. TET-Mediated Sequestration of miR-26 Drives EZH2 Expression and Gastric Carcinogenesis. Cancer Res. 2017, 77, 6069–6082. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.M.; Cha, J.Y.; Kim, J.; Kim, D.; Trang, H.T.H.; Kim, Y.M.; Cho, Y.H.; Park, D.; Hong, S. Smad3 regulates E-cadherin via miRNA-200 pathway. Oncogene 2012, 31, 3051–3059. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Deng, M.; Tang, Y.; Xie, X.; Guo, J.; Kong, Y.; Ye, F.; Su, Q.; Xie, X. miR-200b and miR-200c as Prognostic Factors and Mediators of Gastric Cancer Cell Progression. Clin. Cancer Res. 2013, 19, 5602–5612. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.S.; Guo, X.W.; Zhang, G.; Liang, L.X.; Nong, B. The Diagnostic and Prognostic Value of miR-200c in Gastric Cancer: A Meta-Analysis. Dis. Markers 2019, 2019, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, X.; Wang, Z.; Fillmore, R.; Xi, Y. MiR-200, a new star miRNA in human cancer. Cancer Lett. 2014, 344, 166–173. [Google Scholar] [CrossRef]
- Chang, L.; Guo, F.; Huo, B.; Lv, Y.; Wang, Y.; Liu, W. Expression and clinical significance of the microRNA-200 family in gastric cancer. Oncol. Lett. 2015, 9, 2317–2324. [Google Scholar] [CrossRef] [Green Version]
- Cong, N.; Du, P.; Zhang, A.; Shen, F.; Su, J.; Pu, P.; Wang, T.; Zjang, J.; Kang, C.; Zhang, Q. Downregulated microRNA-200a promotes EMT and tumor growth through the Wnt/β-catenin pathway by targeting the E-cadherin repressors ZEB1/ZEB2 in gastric adenocarcinoma. Oncol. Rep. 2013, 29, 1579–1587. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]


| Parameter | Total | |
|---|---|---|
| n = 33 | % | |
| Sex | ||
| F | 13 | 39.4 |
| M | 20 | 60.6 |
| Age, years | ||
| <77 | 15 | 45.5 |
| ≥77 | 18 | 54.5 |
| T | ||
| 1 | 2 | 6.1 |
| 2 | 12 | 36.4 |
| 3 | 11 | 33.3 |
| 4 | 6 | 18.2 |
| N/A | 2 | 6.1 |
| N | ||
| 0 | 13 | 39.4 |
| + | 18 | 54.5 |
| N/A | 2 | 6.1 |
| M | ||
| 0 | 11 | 33.3 |
| 1 | 2 | 6.1 |
| X | 7 | 21.2 |
| N/A | 13 | 39.4 |
| Grade | ||
| 1 | 1 | 3.0 |
| 2 | 8 | 24.2 |
| 3 | 17 | 51.5 |
| N/A | 7 | 21.2 |
| Tumor site | ||
| Cardia | 3 | 9.1 |
| Fundus | 1 | 3.0 |
| Body | 8 | 24.2 |
| Antrum | 10 | 30.3 |
| N/A | 11 | 33.3 |
| Tumor size (cm) | ||
| <5 | 16 | 48.9 |
| ≥5 | 17 | 51.1 |
| Helicobacter pylori | ||
| Positive | 16 | 48.5 |
| Negative | 14 | 42.4 |
| N/A | 3 | 9.1 |
| miR | In Silico Prediction | Experimental Evidence | |||
|---|---|---|---|---|---|
| Target Gene | miRTarBase | miRDB | TargetScan | miR-Target Interaction (+) or miR Expression Deregulation [ref.] in GC | |
| miR-506 | SNAI2 | + | + | + | + |
| miR-141 | ZEB1 | + | + | + | + |
| ZEB2 | + | + | + | + | |
| miR-217 | EZH2 | + | + | + | + |
| miR-429 | EZH2 | + | + | + | + |
| miR-199a | CDH1 | + | + | + | + |
| miR-200a | ZEB1 | + | + | + | + |
| ZEB2 | + | + | + | + | |
| miR-200b | ZEB1 | + | + | + | + |
| ZEB2 | + | + | + | + | |
| miR-200c | ZEB1 | + | + | + | + |
| ZEB2 | + | + | + | + | |
| SUZ12 | + | + | + | + | |
| miR-101 | EZH2 | + | + | + | + |
| ZEB1 | + | + | + | + | |
| miR-153 | SNAI1 | + | + | + | + |
| miR-26b | EZH2 | + | + | + | [15,16] |
| miR-23a | CDH1 | + | + | + | + |
| miR-544 | CDH1 | + | + | + | + |
| miR-34c | SNAI1 | + | + | + | [17] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossi, T.; Tedaldi, G.; Petracci, E.; Abou Khouzam, R.; Ranzani, G.N.; Morgagni, P.; Saragoni, L.; Monti, M.; Calistri, D.; Ulivi, P.; et al. E-cadherin Downregulation and microRNAs in Sporadic Intestinal-Type Gastric Cancer. Int. J. Mol. Sci. 2019, 20, 4452. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20184452
Rossi T, Tedaldi G, Petracci E, Abou Khouzam R, Ranzani GN, Morgagni P, Saragoni L, Monti M, Calistri D, Ulivi P, et al. E-cadherin Downregulation and microRNAs in Sporadic Intestinal-Type Gastric Cancer. International Journal of Molecular Sciences. 2019; 20(18):4452. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20184452
Chicago/Turabian StyleRossi, Tania, Gianluca Tedaldi, Elisabetta Petracci, Raefa Abou Khouzam, Guglielmina Nadia Ranzani, Paolo Morgagni, Luca Saragoni, Manlio Monti, Daniele Calistri, Paola Ulivi, and et al. 2019. "E-cadherin Downregulation and microRNAs in Sporadic Intestinal-Type Gastric Cancer" International Journal of Molecular Sciences 20, no. 18: 4452. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20184452






