Glycan Binding Profiling of Jacalin-Related Lectins from the Pteria Penguin Pearl Shell
Abstract
:1. Introduction
2. Results
2.1. Purification and Characterization of Novel Lectins from the Secretory Fluid of Pteria Penguin Mantle
2.2. Amino Acid Sequences of PPL3 and PPL4 Subunits
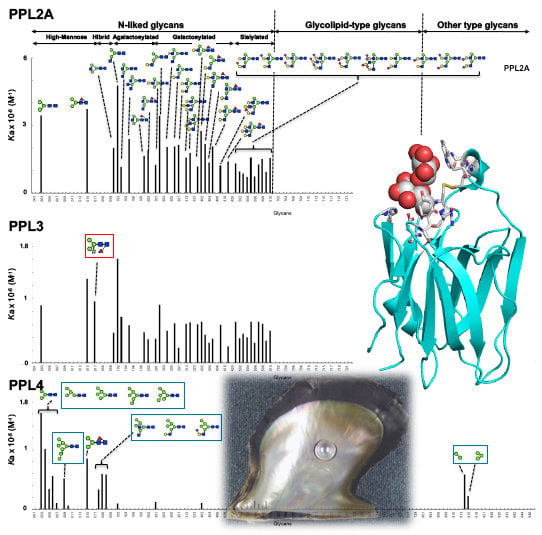
2.3. Carbohydrate-Binding Profilings of PPL2A, PPL3s, and PPL4
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Isolation of Lectins from the Secretory Fluid of Mantle
4.3. Amino Acid Sequences of PPL3 and PPL4 Subunits
4.4. Sugar-Binding Specificity
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CAM | S-carboxamidomethylated |
| CCA | Cancer antennarius lectin |
| CRD | carbohydrate recognition domain |
| FAC | frontal affinity chromatography |
| GlcNAc | N-acetyl-D-glucosamine |
| HPLC | high-performance liquid chromate |
| JRL | jacalin-related lectin |
| PPL | Pteria penguin lectin |
| SDS-PAGE | sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| SEM | scanning electron microscopy |
Appendix A
References
- Zhang, C.; Zhang, R. Matrix proteins in the outer shells of mollusks. Mar. Biotechnol. 2013, 8, 572–586. [Google Scholar] [CrossRef] [PubMed]
- Joubert, C.; Piquemal, D.; Marie, B.; Manchon, L.; Pierrat, F.; Zanella-Cléon, I.; Cochennec-Laureau, N.; Gueguen, Y.; Montagnani, C. Transcriptome and proteome analysis of Pinctada margaritifera calcifying mantle and shell: Focus on biomineralization. BMC Genom. 2010, 11, 613. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, S.; Wang, N.; Inoue, H.; Maeyama, K.; Okamoto, K.; Nagai, K.; Kondo, H.; Hirono, I.; Asakawa, S.; Watabe, S. Deep sequencing of ESTs from nacreous and prismatic layer producing tissues and a screen for novel shell formation-related genes in the pearl oyster. PLoS ONE 2011, 6, e21238. [Google Scholar] [CrossRef] [PubMed]
- Fang, D.; Xu, G.; Hu, Y.; Pan, C.; Zhang, R. Identification of genes directly involved in shell formation and their functions in pearl oyster, Pinctada fucata. PLoS ONE 2011, 6, e21860. [Google Scholar] [CrossRef] [PubMed]
- Marie, B.; Joubert, C.; Tayale, A.; Zanella-Cleon, I.; Belliard, C.; Piquemal, D.; Cochennec-Laureau, N.; Marin, F.; Gueguen, Y.; Montagnani, C. Different secretory repertoires control the biomineralization processes of prism and nacre deposition of the pearl oyster shell. Proc. Natl. Acad. Sci. USA 2012, 109, 20986–20991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeuchi, T.; Kawashima, T.; Koyanagi, R.; Gyoja, F.; Tanaka, M.; Ikuta, T.; Shoguchi, E.; Fujiwara, M.; Shinzato, C.; Hisata, K.; et al. Draft genome of the pearl oyster Pinctada fucata: A platform for understanding bivalve biology. DNA Res. 2012, 19, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, H.; Endo, H.; Hashimoto, N.; Iimura, K.; Isowa, Y.; Kinoshita, S.; Kotaki, T.; Masaoka, T.; Miki, T.; Nakayama, S.; et al. The Diversity of shell matrix proteins: Genome-wide investigation of the pearl oyster, Pinctada fucata. Zool. Sci. 2013, 30, 801–816. [Google Scholar] [CrossRef]
- Freer, A.; Bridgett, S.; Jiang, J.; Cusack, M. Biomineral Proteins from Mytilus edulis Mantle Tissue Transcriptome. Mar. Biotechnol. 2014, 16, 34–45. [Google Scholar] [CrossRef]
- Zhang, D.; Jiang, S.; Hu, Y.; Cui, S.; Guo, H.; Wu, K.; Li, Y.; Su, T. A multidomain galectin involved in innate immune response of pearl oyster Pinctada fucata. Dev. Comp. Immunol. 2011, 35, 1–6. [Google Scholar] [CrossRef]
- Anju, A.; Jeswin, J.; Thomas, P.C.; Vijayan, K.K. Molecular cloning, characterization and expression analysis of F-type lectin from pearl oyster Pinctada fucata. Fish Shellfish Immunol. 2013, 35, 170–174. [Google Scholar] [CrossRef]
- Multigner, L.; Decaro, A.; Lombardo, D.; Campese, D.; Sarles, H. Pancreatic stone protein, a phosphoprotein which inhibits calcium carbonate precipitation from human pancreatic juice. Biochem. Biophys. Res. Commun. 1983, 110, 69–74. [Google Scholar] [CrossRef]
- Muramoto, K.; Yako, H.; Murakami, K.; Odo, S.; Kamiya, H. Inhibition of the growth of calcium-carbonate crystals by multiple lectins in the celomic fluid of the acorn barnacle Megabalanus-rosa. Comp. Biochem. Phys. B Biochem. Mol. Biol. 1994, 107, 401–409. [Google Scholar] [CrossRef]
- Mann, K.; Siedler, F. The amino acid sequence of ovocleidin 17, a major protein of the avian eggshell calcified layer. Biochem. Mol. Biol. Int. 1999, 47, 997–1007. [Google Scholar] [PubMed]
- Mann, K.; Weiss, I.M.; Andre, S.; Gabius, H.J.; Fritz, M. The amino-acid sequence of the abalone (Haliotis laevigata) nacre protein perlucin-Detection of a functional C-type lectin domain with galactose/mannose specificity. Eur. J. Biochem. 2000, 267, 5257–5264. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.L.; Gonzales, K.; SEaver, R.W.; Livingston, B.T. The skeletal proteome of the brittle star Ophiothrix spiculata identifies C-type lectins and other proteins conserved in echinoderm skeleton formation. AIMS Mol. Sci. 2016, 3, 357–367. [Google Scholar] [CrossRef]
- Flores, R.L.; Livingston, B.T. The skeletal proteome of the sea star Patiria miniata and evolution of biomineralization in echinoderms. BMC Evol. Biol. 2017, 17, 125. [Google Scholar] [CrossRef] [PubMed]
- Blank, S.; Arnoldi, M.; Khoshnavaz, S.; Treccani, L.; Kuntz, M.; Mann, K.; Grathwohl, G.; Fritz, M. The nacre protein perlucin nucleates growth of calcium carbonate crystals. J. Microsc. 2003, 212, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Weiss, I.M.; Kaufmann, S.; Mann, K.; Fritz, M. Purification and characterization of perlucin and perlustrin, two new proteins from the shell of the mollusk Haliotis laevigata. Biochem. Biophys. Res. Commun. 2000, 267, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Dodenhof, T.; Dietz, F.; Franken, S.; Grunwald, I.; Kelm, S. Splice variants of perlucin from Haliotis laevigata modulate the crystallization of CaCO3. PLoS ONE 2014, 9, e97126. [Google Scholar] [CrossRef] [PubMed]
- Livingston, B.T.; Killian, C.E.; Wilt, F.; Cameron, A.; Landrum, M.J.; Ermolaeva, O.; Sapojnikov, V.; Maglott, D.R.; Buchanan, A.M.; Ettensohn, C.A. A genome-wide analysis of biomineralization-related proteins in the sea urchin Strongylocentrotus purpuratus. Dev. Biol. 2006, 300, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Liener, I.E.; Sharon, N.; Goldstein, I.J. The Lectins Properties, Functions, and Applications in Biology and Medicine; Academic Press, Elsevier Ltd.: Amsterdam, Netherlands, 1986; p. 600. [Google Scholar]
- Naganuma, T.; Ogawa, T.; Hirabayashi, J.; Kasai, K.; Kamiya, H.; Muramoto, K. Isolation, characterization and molecular evolution of a novel pearl shell lectin from a marine bivalve, Pteria penguin. Mol. Divers. 2006, 10, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Naganuma, T.; Hoshino, W.; Shikanai, Y.; Sato, R.; Liu, K.; Sato, S.; Muramoto, K.; Osada, M.; Yoshimi, K.; Ogawa, T. Novel matrix proteins of Pteria penguin pearl oyster shell nacre homologous to the Jacalin-related β-prism fold lectins. PLoS ONE 2014, 9, e112326. [Google Scholar] [CrossRef] [PubMed]
- Nakae, S.; Shionyu, M.; Ogawa, T.; Shirai, T. Structures of jacalin-related lectin PPL3 regulating pearl shell biomineralization. Proteins 2018, 86, 644–653. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System. ver. 2.3.2. 2002. Available online: http://www.pymol.org (accessed on 5 September 2019).
- Hirabayashi, J.; Arata, Y.; Kasai, K. Frontal Affinity Chromatography as a Tool for Elucidation of Sugar Recognition Properties of Lectins. Methods Enzymol. 2003, 362, 353–368. [Google Scholar] [PubMed]
- Nakamura-Tsuruta, S.; Uchiyama, N.; Hirabayashi, J. High-throughput analysis of lectin-oligosaccharide interactions by automated frontal affinity chromatography. Methods Enzymol. 2006, 415, 311–325. [Google Scholar]
- Kimura, Y.; Ohno, A.; Takagi, S. Structural analysis of N-glycans of storage glycoproteins in soybean (Glycine max. L) seed. Biosci. Biotechnol. Biochem. 1997, 61, 1866–1871. [Google Scholar] [CrossRef]
- Bourne, Y.; Roig-Zamboni, V.; Barre, A.; Peumans, W.J.; Astoul, C.H.; Van Damme, E.J.M.; Rougé, P. The crystal structure of the calystegia sepium agglutinin reveals a novel quaternary arrangement of lectin subunits with a β-prism fold. J. Biol. Chem. 2004, 279, 527–533. [Google Scholar] [CrossRef]
- Meagher, J.L.; Winter, H.C.; Porscha, E.; Goldstein, I.J.; Stuckey, J.A. Crystal structure of banana lectin reveals a novel second sugar binding site. Glycobiology 2005, 15, 1033–1042. [Google Scholar] [CrossRef]
- Kanagawa, M.; Satoh, T.; Ikeda, A.; Nakano, Y.; Yagi, H.; Katoc, K.; Kojima-Aikawa, K.; Yamaguchi, Y. Crystal structures of human secretory proteins ZG16p and ZG16b reveal a Jacalin-related β-prism fold. Biochem. Biophys. Res. Commun. 2011, 404, 201–205. [Google Scholar] [CrossRef]
- Nakamura-Tsuruta, S.; Uchiyama, N.; Peumans, W.J.; Van Damme, E.J.M.; Totani, K.; Ito, Y.; Hirabayashi, J. Analysis of the sugar-binding specificity of mannose-binding-type Jacalin-related lectins by frontal affinity chromatography–an approach to functional classification. FEBS J. 2008, 275, 1227–1239. [Google Scholar] [CrossRef] [PubMed]
- Weiner, S.; Traub, W. X-ray diffraction study of the insoluble organic matrix of mollusk shells. FEBS Lett. 1980, 111, 311–316. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, M.; Sakuda, S.; Nagasawa, H. Identification of chitin in the prismatic layer of the shell and a chitin synthase gene from the Japanese pearl oyster, Pinctada fucata. Biosci. Biotechnol. Biochem. 2007, 71, 1735–1744. [Google Scholar] [CrossRef] [PubMed]
- Weiss, I.M.; Schönitzer, V.; Eichner, N.; Sumper, M. The chitin synthase involved in marine bivalve mollusk shell formation contains a myosin domain. FEBS Lett. 2006, 580, 1846–1852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiss, I.M.; Lüke, F.; Eichner, N.; Guth, C.; Clausen-Schaumann, H. On the function of chitin synthase extracellular domains in biomineralization. J. Struct. Biol. 2013, 183, 216–225. [Google Scholar] [CrossRef]
- Levi-Kalisman, Y.; Falini, G.; Addadi, L.; Weiner, S. Structure of the nacreous organic matrix of a bivalve mollusk shell examined in the hydrated state using Cryo-TEM. J. Struct. Biol. 2001, 135, 8–17. [Google Scholar] [CrossRef]
- Arias, J.L.; Fernández, M.S. Polysaccharides and proteoglycans in calcium carbonate-based biomineralization. Chem. Rev. 2008, 108, 4475–4482. [Google Scholar] [CrossRef]
- Stone, K.L.; McNulty, D.E.; LoPresti, M.L.; Crawford, J.M.; DeAngelis, R.; Williams, K.R. Elution and internal amino acid sequencing of PVDF blotted proteins. In Techniques in Protein Chemistry III; Angeletti, R.M., Ed.; Academic Press: San Diego, CA, USA, 1992; pp. 23–34. [Google Scholar]






| Saccharides/Glycoproteins | PPL2A a | PPL2B a | PPL3 | PPL4 | |
|---|---|---|---|---|---|
| mM b | |||||
| D-Glucose | >250 | >250 | 200 | 25 | |
| D-Fructose | 250 | >250 | 50 | 50 | |
| D-Mannose | >250 | >250 | 100 | 25 | |
| D-Glucosamine | >250 | >250 | >200 | 200 | |
| N-Acetyl-D-glucosamine | 250 | >250 | >200 | 6.25 | |
| Trehalose | Glc(α1-1α)Glc | 7.8 | >250 | 25 | 100 |
| Kojibiose | Glc(α1-2α)Glc | >250 | 125 | >200 | 100 |
| Maltose | Glc(α1-4α)Glc | >250 | >250 | >200 | 200 |
| Isomaltose | Glc(α1-6α)Glc | 7.8 | >250 | 12.5 | >200 |
| Others c | >250 | >250 | >200 | >200 | |
| %(w/v) b | |||||
| Chitin oligomers | GlcNAc(β1-4β)GlcNAc | >0.25 | >0.25 | >0.25 | 0.125 |
| Chitosan oligomers | GlcNH2(β1-4β)GlcNH2 | >0.25 | >0.25 | >0.25 | >0.25 |
| Fetuin | >0.25 | 0.25 | 0.025 | 0.025 | |
| Asialo fetuin | 0.078 | 0.078 | 0.0125 | 0.025 | |
| Other d | >0.25 | >0.25 | >0.25 | >0.25 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ogawa, T.; Sato, R.; Naganuma, T.; Liu, K.; Lakudzala, A.E.; Muramoto, K.; Osada, M.; Yoshimi, K.; Hiemori, K.; Hirabayashi, J.; et al. Glycan Binding Profiling of Jacalin-Related Lectins from the Pteria Penguin Pearl Shell. Int. J. Mol. Sci. 2019, 20, 4629. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20184629
Ogawa T, Sato R, Naganuma T, Liu K, Lakudzala AE, Muramoto K, Osada M, Yoshimi K, Hiemori K, Hirabayashi J, et al. Glycan Binding Profiling of Jacalin-Related Lectins from the Pteria Penguin Pearl Shell. International Journal of Molecular Sciences. 2019; 20(18):4629. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20184629
Chicago/Turabian StyleOgawa, Tomohisa, Rie Sato, Takako Naganuma, Kayeu Liu, Agness Ethel Lakudzala, Koji Muramoto, Makoto Osada, Kyosuke Yoshimi, Keiko Hiemori, Jun Hirabayashi, and et al. 2019. "Glycan Binding Profiling of Jacalin-Related Lectins from the Pteria Penguin Pearl Shell" International Journal of Molecular Sciences 20, no. 18: 4629. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20184629