Molecular Modeling Studies on the Binding Mode of the PD-1/PD-L1 Complex Inhibitors
Abstract
:1. Introduction
2. Results and Discussion
2.1. Docking Scores and Validation
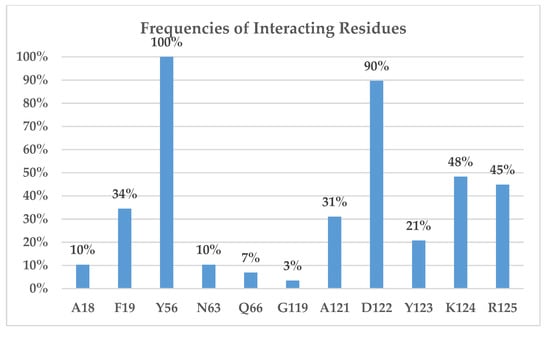
2.2. Binding Interactions of PD-L1/inhibitors
3. Computational Methods
3.1. Preparation of Protein Structures
3.2. Preparation of Ligands and Molecular Docking
3.3. Binding Free Energy Calculations Using the Moveable-Type (MT)-Based Approach
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharpe, A.H.; Wherry, E.J.; Ahmed, R.; Freeman, G.J. The function of programmed cell death 1 and its ligands in regulating autoimmunity and infection. Nat. Immunol. 2007, 8, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Nirschl, C.J.; Drake, C.G. Molecular pathways: Coexpression of immune checkpoint molecules: Signaling pathways and implications for cancer immunotherapy. Clin. Cancer Res. 2013, 19, 4917–4924. [Google Scholar] [CrossRef] [PubMed]
- Mahoney, K.M.; Rennert, P.D.; Freeman, G.J. Combination cancer immunotherapy and new immunomodulatory targets. Nat. Rev. Drug Discov. 2015, 14, 561–584. [Google Scholar] [CrossRef] [PubMed]
- Topalian, S.L.; Drake, C.G.; Pardoll, D.M. Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer Cell. 2015, 27, 450–461. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Allison, J.P. The future of immune checkpoint therapy. Science. 2015, 348, 56–61. [Google Scholar] [CrossRef]
- Shin, D.S.; Ribas, A. The evolution of checkpoint blockade as a cancer therapy: What’s here, what’s next? Curr. Opin. Immunol. 2015, 33, 23–35. [Google Scholar] [CrossRef]
- Suh, W.; Gajewska, B.U.; Okada, H.; Gronski, M.A.; Bertram, E.M.; Dawicki, W.; Duncan, G.S.; Bukczynski, J.; Plyte, S.; Elia, A.; et al. The B7 family member B7-H3 preferentially down-regulates T helper type 1-mediated immune responses. Nat. Immunol. 2003, 4, 899–906. [Google Scholar] [CrossRef]
- Greenwald, R.J.; Freeman, G.J.; Sharpe, A.H. The B7 family revisited. Annu. Rev. Immunol. 2005, 23, 515–548. [Google Scholar] [CrossRef]
- Parry, R.V.; Chemnitz, J.M.; Frauwirth, K.A.; Lanfranco, A.R.; Braunstein, I.; Kobayashi, S.V.; Linsley, P.S.; Thompson, C.B.; Riley, J.L. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol. Cell Biol. 2005, 25, 9543–9553. [Google Scholar] [CrossRef]
- Sharma, P.; Allison, J.P. Immune checkpoint targeting in cancer therapy: Toward combination strategies with curative potential. Cell. 2015, 161, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Hoos, A. Development of immuno-oncology drugs—From CTLA4 to PD1 to the next generations. Nat. Rev. Drug Discov. 2016, 15, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Lipson, E.J.; Drake, C.G. Ipilimumab: An anti-CTLA-4 antibody for metastatic melanoma. Clin. Cancer Res. 2011, 17, 6958–6962. [Google Scholar] [CrossRef] [PubMed]
- Dömling, A.; Holak, T.A. Programmed death-1: Therapeutic success after more than 100 years of cancer immunotherapy. Angew Chem. Int. Ed. Engl. 2014, 53, 2286–2288. [Google Scholar] [CrossRef] [PubMed]
- Powles, T.; Eder, J.P.; Fine, G.D.; Braiteh, F.S.; Loriot, Y.; Cruz, C.; Bellmunt, J.; Burris, H.A.; Petrylak, D.P.; Teng, S.L.; et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature 2014, 515, 558–562. [Google Scholar] [CrossRef] [PubMed]
- Hamid, O.; Robert, C.; Daud, A.; Hodi, F.S.; Hwu, W.J.; Kefford, R.; Wolchok, J.D.; Hersey, P.; Joseph, R.W.; Weber, J.S.; et al. Safety and tumor responses with lambrolizumab (anti-PD-1) in melanoma. N. Engl. J. Med. 2013, 369, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Brahmer, J.R.; Tykodi, S.S.; Chow, L.Q.M.; Hwu, W.J.; Topalian, S.L.; Hwu, P.; Drake, C.G.; Camacho, L.H.; Kauh, J.; Odunsi, K.; et al. Safety and activity of Anti–PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 2012, 366, 2455–2465. [Google Scholar] [CrossRef]
- Keir, M.E.; Butte, M.J.; Freeman, G.J.; Sharpe, A.H. PD-1 and its ligands in tolerance and immunity. Annu. Rev. Immunol. 2008, 26, 677–704. [Google Scholar] [CrossRef] [PubMed]
- Francisco, L.M.; Sage, P.T.; Sharpe, A.H. The PD-1 pathway in tolerance and autoimmunity. Immunol. Rev. 2010, 236, 219–242. [Google Scholar] [CrossRef]
- Okazaki, T.; Honjo, T. PD-1 and PD-1 ligands: From discovery to clinical application. Int. Immunol. 2007, 19, 813–824. [Google Scholar] [CrossRef]
- Liang, S.C.; Latchman, Y.E.; Buhlmann, J.E.; Tomczak, M.F.; Horwitz, B.H.; Freeman, G.J.; Sharpe, A.H. Regulation of PD-1, PD-L1, and PD-L2 expression during normal and autoimmune responses. Eur. J. Immunol. 2003, 33, 2706–2716. [Google Scholar] [CrossRef]
- Nishimura, H.; Nose, M.; Hiai, H.; Minato, N.; Honjo, T. Development of lupus-like autoimmune diseases by disruption of the PD-1 gene encoding an ITIM motif-carrying immunoreceptor. Immunity 1999, 11, 141–151. [Google Scholar] [CrossRef]
- Ansari, M.J.I.; Salama, A.D.; Chitnis, T.; Smith, R.N.; Yagita, H.; Akiba, H.; Yamazaki, T.; Azuma, M.; Iwai, H.; Khoury, S.J.; et al. The programmed death-1 (PD-1) pathway regulates autoimmune diabetes in nonobese diabetic (NOD) mice. J. Exp. Med. 2003, 198, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Keir, M.E.; Liang, S.C.; Guleria, I.; Latchman, Y.E.; Qipo, A.; Albacker, L.A.; Koulmanda, M.; Freeman, G.J.; Sayegh, M.H.; Sharpe, A.H. Tissue expression of PD-L1 mediates peripheral T cell tolerance. J. Exp. Med. 2006, 203, 883–895. [Google Scholar] [CrossRef] [PubMed]
- Phan, T.G.; Long, G.V.; Scolyer, R.A. Checkpoint inhibitors for cancer immunotherapy. Multiple checkpoints on the long road towards cancer immunotherapy. Immunol. Cell Biol. 2015, 93, 323–325. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Soria, J.; Kowanetz, M.; Fine, G.D.; Hamid, O.; Gordon, M.S.; Sosman, J.A.; McDermott, D.F.; Powderly, J.D.; Gettinger, S.N.; et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014, 515, 563–567. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wherry, E.J. T cell exhaustion. Nature Immunol. 2011, 12, 492–499. [Google Scholar] [CrossRef]
- Sakuishi, K.; Apetoh, L.; Sullivan, J.M.; Blazar, B.R.; Kuchroo, V.K.; Anderson, A.C. Targeting tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J. Exp. Med. 2010, 207, 2187–2194. [Google Scholar] [CrossRef]
- Sun, S.; Fei, X.; Mao, Y.; Wang, X.; Garfield, D.H.; Huang, O.; Wang, J.; Yuan, F.; Sun, L.; Yu, Q.; et al. PD-1(+) immune cell infiltration inversely correlates with survival of operable breast cancer patients. Cancer Immunol. Immunother. 2014, 63, 395–406. [Google Scholar] [CrossRef]
- Muenst, S.; Soysal, S.D.; Gao, F.; Obermann, E.C.; Oertli, D.; Gillanders, W.E. The presence of programmed death 1 (PD-1)-positive tumor-infiltrating lymphocytes is associated with poor prognosis in human breast cancer. Breast Cancer Res. Treat. 2013, 139, 667–676. [Google Scholar] [CrossRef]
- Ahmadzadeh, M.; Johnson, L.A.; Heemskerk, B.; Wunderlich, J.R.; Dudley, M.E.; White, D.E.; Rosenberg, S.A. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood 2009, 114, 1537–1544. [Google Scholar] [CrossRef]
- Matsuzaki, J.; Gnjatic, S.; Mhawech-Fauceglia, P.; Beck, A.; Miller, A.; Tsuji, T.; Eppolito, C.; Qian, F.; Lele, S.; Shrikant, P.; et al. Tumor-infiltrating NY-ESO-1–specific CD8 T cells are negatively regulated by LAG-3 and PD-1 in human ovarian cancer. Proc. Natl. Acad. Sci. USA. 2010, 107, 7875–7880. [Google Scholar] [CrossRef] [PubMed]
- Hawkes, E.A.; Grigg, A.; Chong, G. Programmed cell death-1 inhibition in lymphoma. Lancet Oncol. 2015, 16, 234. [Google Scholar] [CrossRef]
- Inman, B.A.; Sebo, T.J.; Frigola, X.; Dong, H.; Bergstralh, E.J.; Frank, I.; Fradet, Y.; Lacombe, L.; Kwon, E.D. PD-L1 (B7-H1) expression by urothelial carcinoma of the bladder and BCG-induced granulomata: Associations with localized stage progression. Cancer 2007, 109, 1499–1505. [Google Scholar] [CrossRef] [PubMed]
- Khalil, D.N.; Smith, E.L.; Brentjens, R.J.; Wolchok, J.D. The future of cancer treatment: Immunomodulation, CARs and combination immunotherapy. Nat. Rev. Clin. Oncol. 2016, 13, 273–290. [Google Scholar] [CrossRef] [PubMed]
- Baumeister, S.H.; Freeman, G.J.; Dranoff, G.; Sharpe, A.H. Coinhibitory pathways in immunotherapy for cancer. Annu. Rev. Immunol. 2016, 34, 539–573. [Google Scholar] [CrossRef] [PubMed]
- Farkona, S.; Diamandis, E.P.; Blasutig, I.M. Cancer immunotherapy: The beginning of the end of cancer? BMC Med. 2016, 14, 73. [Google Scholar] [CrossRef] [PubMed]
- Kruger, S.; Ilmer, M.; Kobold, S.; Cadilha, B.L.; Endres, S.; Ormanns, S.; Cadilha, B.L.; Endres, S.; Ormanns, S.; Schuebbe, G.; et al. Advances in cancer immunotherapy 2019—Latest trends. J. Exp. Clin. Cancer Res. 2019, 38, 268. [Google Scholar] [CrossRef] [PubMed]
- Danlos, F.X.; Voisin, A.L.; Dyevre, V.; Michot, J.M.; Routier, E.; Taillade, L.; Champiat, S.; Aspeslagh, S.; Haroche, J.; Albiges, L.; et al. Safety and efficacy of anti-programmed death 1 antibodies in patients with cancer and pre-existing autoimmune or inflammatory disease. Eur. J. Cancer 2018, 91, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, R.; Mezquita, L.; Texier, M.; Lahmar, J.; Audigier-Valette, C.; Tessonnier, L.; Mazieres, J.; Zalcman, G.; Brosseau, S.; Le Moulec, S.; et al. Hyperprogressive Disease in Patients with Advanced Non-Small Cell Lung Cancer Treated with PD-1/PD-L1 Inhibitors or with Single-Agent Chemotherapy. JAMA Oncol. 2018, 4, 1543–1552. [Google Scholar] [CrossRef] [PubMed]
- Lecis, D.; Sangaletti, S.; Colombo, M.P.; Chiodoni, C. Immune Checkpoint Ligand Reverse Signaling: Looking Back to Go Forward in Cancer Therapy. Cancers (Basel) 2019, 11, 624. [Google Scholar] [CrossRef] [PubMed]
- Zak, K.M.; Kitel, R.; Przetocka, S.; Golik, P.; Guzik, K.; Musielak, B.; Dömling, A.; Dubin, G.; Holak, T.A. Structure of the complex of human programmed death 1, PD-1, and its ligand PD-L. Structure 2015, 23, 2341–2348. [Google Scholar] [CrossRef]
- Lee, H.T.; Lee, J.Y.; Lim, H.; Lee, S.H.; Moon, Y.J.; Pyo, H.J.; Ryu, S.E.; Shin, W.; Heo, Y.S. Molecular mechanism of PD-1/PD-L1 blockade via anti-PD-L1 antibodies atezolizumab and durvalumab. Sci. Rep. 2017, 7, 5532. [Google Scholar] [CrossRef] [PubMed]
- Chupak, L.S.; Zheng, X. Compounds Useful as Immunomodulators. WO 2015034820 A1, 12 March 2015. [Google Scholar]
- Chupak, L.S.; Ding, M.; Martin, S.W.; Zheng, X.; Hewawasam, P.; Connolly, T.P.; Xu, N.; Yeung, K.A.; Zhu, J.; Langley, D.R.; et al. Compounds Useful as Immunomodulators. WO 2015160641 A2, 22 October 2015. [Google Scholar]
- Zak, K.M.; Grudnik, P.; Guzik, K.; Zieba, B.J.; Musielak, B.; Dömling, A.; Dubin, G.; Holak, T.A. Structural basis for small molecule targeting of the programmed death ligand 1 (PD-L1). Oncotarget. 2016, 7, 30323–30335. [Google Scholar] [CrossRef] [PubMed]
- Guzik, K.; Zak, K.M.; Grudnik, P.; Magiera, K.; Musielak, B.; Törner, R.; Skalniak, L.; Dömling, A.; Dubin, G.; Holak, T.A. Small-molecule inhibitors of the programmed cell death-1/programmed death-ligand 1 (PD-1/PD-L1) interaction via transiently induced protein states and dimerization of PD-L. J. Med. Chem. 2017, 60, 5857–5867. [Google Scholar] [CrossRef] [PubMed]
- Skalniak, L.; Zak, K.M.; Guzik, K.; Magiera, K.; Musielak, B.; Pachota, M.; Szelazek, B.; Kocik, J.; Grudnik, P.; Tomala, M.; et al. Small-molecule inhibitors of PD-1/PD-L1 immune checkpoint alleviate the PD-L1-induced exhaustion of T-cells. Oncotarget. 2017, 8, 72167–72181. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Merz, K.M., Jr. Development of the knowledge-based and empirical combined scoring algorithm (KECSA) to score protein–ligand interactions. J. Chem. Inf. Model. 2013, 53, 1073–1083. [Google Scholar] [CrossRef]
- Zhong, H.A.; Santos, E.M.; Vasileiou, C.; Zheng, Z.; Geiger, J.H.; Borhan, B.; Merz, K.M., Jr. Free-Energy-Based Protein Design: Re-Engineering Cellular Retinoic Acid Binding Protein II Assisted by the Moveable-Type Approach. J. Am. Chem. Soc. 2018, 140, 3483–3486. [Google Scholar] [CrossRef]
- Cole, J.C.; Murray, C.W.; Nissink, J.W.M.; Taylor, R.D.; Taylor, R. Comparing protein–ligand docking programs is difficult. Proteins 2005, 60, 325–332. [Google Scholar] [CrossRef]
- Jain, A.N. Bias, reporting, and sharing: Computational evaluations of docking methods. J. Comput. Aided Mol. Des. 2008, 22, 201–212. [Google Scholar] [CrossRef]
- Hevener, K.E.; Zhao, W.; Ball, D.M.; Babaoglu, K.; Qi, J.; White, S.W.; Lee, R.E. Validation of molecular docking programs for virtual screening against dihydropteroate synthase. J. Chem. Inf. Model. 2009, 49, 444–460. [Google Scholar] [CrossRef]
- Bender, A.; Glen, R.C. A discussion of measures of enrichment in virtual screening: Comparing the information content of descriptors with increasing levels of sophistication. J. Chem. Inf. Model. 2005, 45, 1369–1375. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Shoichet, B.K.; Irwin, J.J. Benchmarking sets for molecular docking. J. Med. Chem. 2006, 49, 6789–6801. [Google Scholar] [CrossRef] [PubMed]
- Almahmoud, S.; Wang, X.; Vennerstrom, J.L.; Zhong, H.A. Conformational Studies of Glucose Transporter 1 (GLUT1) as an Anticancer Drug Target. Molecules 2019, 24, 2159. [Google Scholar] [CrossRef] [PubMed]
- The Molecular Operating Environment (MOE); Chemical Computing Group Inc.: Montreal, QC, Canada, 2019; Available online: https://www.chemcomp.com/Products.htm (accessed on 17 September 2019).
- Case, D.A.; Babin, V.; Berryman, J.T.; Betz, R.M.; Cai, Q.; Cerutti, D.S.; Cheatham Iii, T.E.; Darden, T.A.; Duke, R.E.; Gohlke, H.; et al. AMBER 14, University of California, San Francisco; University of California: San Francisco, CA, USA, 2014. [Google Scholar] [CrossRef]
- Schrödinger, LLC, New York, NY, Schrödinger Suite 2019-1 Protein Preparation wizard, Maestro, Protein Grid Generation, Glide, Macromodel, and Epik. Available online: https://www.schrodinger.com/citations (accessed on 17 September 2019).
- Halgren, T.A. Merck molecular forcefield. I. basis, form, scope, parameterization, and performance of MMFF. J. Comput. Chem. 1996, 17, 490–519. [Google Scholar] [CrossRef]
- NCI Open Database Compounds, Release 3. National Cancer Institute, National Institutes of Health: Bethseda, MD, USA, Septemper 2003. Available online: Http://Cactus.nci.nih.gov/download/nci (accessed on 18 August 2008).
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Sabbah, D.A.; Vennerstrom, J.L.; Zhong, H. Docking studies on isoform-specific inhibition of phosphoinositide-3-kinases. J. Chem. Inf. Model. 2010, 50, 1887–1898. [Google Scholar] [CrossRef] [PubMed]









| Title | IC50 (nM) | Chain C | Chain D |
|---|---|---|---|
| BMS-1001(1, 5NIU) | 2.25 | Tyr56, Asp122, Lys124, Arg125, Phe19 | |
| BMS-200 (2, 5N2F) | 80 | Tyr56 | Tyr56, Ala121, Asp122 |
| BMS-3029 (3) | 2350 | Tyr56, Gln66 | Tyr56, Asp122, Tyr123, Lys124 |
| BMS-1166 (4, 5NIX) | 1.4 | Tyr56, Asp122, Arg125 | |
| BMS-114 (5) | 43 | Tyr56 | Tyr56, Asp122, Arg125 |
| BMS-1197 (6) | 1.85 | Tyr56 | Tyr56, Asp122, Lys124, Arg125, Phe19 |
| BMS-1205 (7) | 2.71 | Tyr56, Gln66 | Tyr56, Asp122, Lys124, Arg125 |
| BMS-1220 (8) | 6.07 | Tyr56, Asp122, Lys124, Arg125 | |
| BMS-2002 (9) | 10 | Tyr56 | Tyr56, Ala121, Asp122, Tyr123, Lys124, Arg125, Phe19 |
| BMS-1250 (10) | 1.19 | Tyr56 | Tyr56, Ala121, Asp122, Arg125, Ala18, Phe19 |
| BMS-1305 (11) | 0.92 | Tyr56 | Tyr56, Asp122, Tyr123, Arg125 |
| BMS-1239 (12) | 148.9 | Tyr56, Asp122, Lys124 | |
| BMS-2010 (13) | 50 | Tyr56, Asp122, Lys124, Arg125, Ala18 | |
| BMS-3024 (14) | 5.54 | Gln66 | Tyr56, Asp122, Arg125, Phe19 |
| BMS-16 (15) | 1945 | Tyr56, Asn63 | Tyr56, Asp122 |
| BMS-82 (16) | 3186 | Tyr56, Ala121, Phe19, Ala18 | |
| BMS-39 (17) | 4184 | Tyr56 | Tyr56, Asp122 |
| BMS-172 (18) | 107 | Tyr56 | Tyr56, Ala121, Asp122, Tyr123 |
| BMS-163 (19) | 93 | Tyr56 | Tyr56, Gly119, Ala121, Asp122, Tyr123 |
| BMS-202 (20, 5J89) | 18 | Tyr56 | Tyr56, Ala121, Asp122 |
| BMS-1043 (21) | 239.2 | Tyr56, Ala121, Asp122, Tyr123, Lys124, Phe19 | |
| BMS-8 (22, 5J8O) | 146 | Asn63 | Tyr56, Lys124 |
| BMS-107 (23) | 329 | Tyr56, Asp122, Lys124 | |
| BMS-101 (24) | 1076 | Gln66 | Tyr56 |
| BMS-1016 (25) | 4.55 | Tyr56 | Tyr56, Asp122, Arg125 |
| BMS-1057 (26) | 985.8 | Tyr56 | Tyr56, Asp122, Lys124, Phe19 |
| BMS-1095 (27) | 81.25 | Tyr56 | Tyr56, Ala121, Asp122, Lys124, Arg125, Phe19 |
| BMS-1108 (28) | 624.2 | Asn63 | Tyr56, Asp122 |
| BMS-1082 (29) | 828.4 | Tyr56, Ala121, Asp122, Lys124, Phe19 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almahmoud, S.; Zhong, H.A. Molecular Modeling Studies on the Binding Mode of the PD-1/PD-L1 Complex Inhibitors. Int. J. Mol. Sci. 2019, 20, 4654. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20184654
Almahmoud S, Zhong HA. Molecular Modeling Studies on the Binding Mode of the PD-1/PD-L1 Complex Inhibitors. International Journal of Molecular Sciences. 2019; 20(18):4654. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20184654
Chicago/Turabian StyleAlmahmoud, Suliman, and Haizhen A. Zhong. 2019. "Molecular Modeling Studies on the Binding Mode of the PD-1/PD-L1 Complex Inhibitors" International Journal of Molecular Sciences 20, no. 18: 4654. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20184654