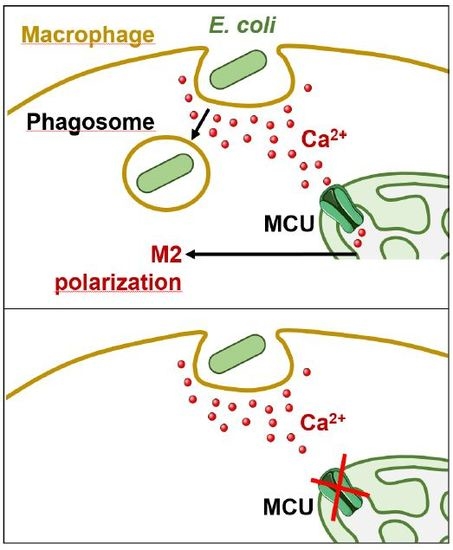
Mitochondrial Calcium Uptake Is Instrumental to Alternative Macrophage Polarization and Phagocytic Activity
Abstract
:1. Introduction
2. Results
2.1. Mitochondrial Calcium Uptake Modulated Macrophage Polarization
2.2. Phagocytosis-Induced Mitochondrial Remodelling and Calcium Uptake
2.3. Blocking MCU Inhibited Macrophage Phagocytosis
3. Discussion
4. Materials and Methods
4.1. Differentiation and Polarization of Human Monocyte-Derived Macrophages
4.2. Gene Expression Analysis
4.3. Flow Cytometry
4.4. Western Blot
4.5. Phagocytosis Assays
4.6. ATP Luminescence Assay
4.7. Gene Silencing
4.8. Imaging Experiments
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Thomas, C.A.; Li, Y.; Kodama, T.; Suzuki, H.; Silverstein, S.C.; El Khoury, J. Protection from lethal gram-positive infection by macrophage scavenger receptor-dependent phagocytosis. J. Exp. Med. 2000, 191, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Donnelly, L.E.; Barnes, P.J. Defective phagocytosis in airways disease. Chest 2012, 141, 1055–1062. [Google Scholar] [CrossRef] [PubMed]
- Sindrilaru, A.; Peters, T.; Schymeinsky, J.; Oreshkova, T.; Wang, H.; Gompf, A.; Mannella, F.; Wlaschek, M.; Sunderkotter, C.; Rudolph, K.L.; et al. Wound healing defect of Vav3−/− mice due to impaired {beta}2-integrin-dependent macrophage phagocytosis of apoptotic neutrophils. Blood 2009, 113, 5266–5276. [Google Scholar] [CrossRef] [PubMed]
- Tabas, I. Macrophage death and defective inflammation resolution in atherosclerosis. Nat. Rev. Immunol. 2010, 10, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Sun, Y.; Liang, C.P.; Thorp, E.B.; Han, S.; Jehle, A.W.; Saraswathi, V.; Pridgen, B.; Kanter, J.E.; Li, R.; et al. Defective phagocytosis of apoptotic cells by macrophages in atherosclerotic lesions of ob/ob mice and reversal by a fish oil diet. Circ. Res. 2009, 105, 1072–1082. [Google Scholar] [CrossRef] [PubMed]
- Bujak, M.; Kweon, H.J.; Chatila, K.; Li, N.; Taffet, G.; Frangogiannis, N.G. Aging-related defects are associated with adverse cardiac remodeling in a mouse model of reperfused myocardial infarction. J. Am. Coll. Cardiol. 2008, 51, 1384–1392. [Google Scholar] [CrossRef] [PubMed]
- Busso, N.; So, A. Mechanisms of inflammation in gout. Arthritis Res. Ther. 2010, 12, 206. [Google Scholar] [CrossRef]
- Alexiewicz, J.M.; Kumar, D.; Smogorzewski, M.; Klin, M.; Massry, S.G. Polymorphonuclear leukocytes in non-insulin-dependent diabetes mellitus: Abnormalities in metabolism and function. Ann. Intern. Med. 1995, 123, 919–924. [Google Scholar] [CrossRef]
- Krol, E.; Agueel, R.; Banue, S.; Smogorzewski, M.; Kumar, D.; Massry, S.G. Amlodipine reverses the elevation in [Ca2+]i and the impairment of phagocytosis in PMNLs of NIDDM patients. Kidney Int. 2003, 64, 2188–2195. [Google Scholar] [CrossRef]
- Nunes, P.; Demaurex, N. The role of calcium signaling in phagocytosis. J. Leukoc. Biol. 2010, 88, 57–68. [Google Scholar] [CrossRef]
- De Stefani, D.; Raffaello, A.; Teardo, E.; Szabo, I.; Rizzuto, R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature 2011, 476, 336–340. [Google Scholar] [CrossRef] [PubMed]
- Marchi, S.; Pinton, P. The mitochondrial calcium uniporter complex: Molecular components, structure and physiopathological implications. J. Physiol. 2014, 592, 829–839. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Yang, W.; Li, J. Ca2+- and protein kinase C-dependent signaling pathway for nuclear factor-kappaB activation, inducible nitric-oxide synthase expression, and tumor necrosis factor-alpha production in lipopolysaccharide-stimulated rat peritoneal macrophages. J. Biol. Chem. 2006, 281, 31337–31347. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Zhang, K.; Wong, D.S.H.; Han, F.; Li, B.; Bian, L. Near-infrared light-controlled regulation of intracellular calcium to modulate macrophage polarization. Biomaterials 2018, 178, 681–696. [Google Scholar] [CrossRef] [PubMed]
- Canton, J. Phagosome maturation in polarized macrophages. J. Leukoc. Biol. 2014, 96, 729–738. [Google Scholar] [CrossRef] [PubMed]
- Tarique, A.A.; Logan, J.; Thomas, E.; Holt, P.G.; Sly, P.D.; Fantino, E. Phenotypic, functional, and plasticity features of classical and alternatively activated human macrophages. Am. J. Respir. Cell. Mol. Biol. 2015, 53, 676–688. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Larson-Casey, J.L.; Carter, A.B. Macrophages utilize the mitochondrial calcium uniporter for profibrotic polarization. FASEB J. 2017, 31, 3072–3083. [Google Scholar] [CrossRef] [Green Version]
- Gu, L.; Larson Casey, J.L.; Andrabi, S.A.; Lee, J.H.; Meza-Perez, S.; Randall, T.D.; Carter, A.B. Mitochondrial calcium uniporter regulates PGC-1alpha expression to mediate metabolic reprogramming in pulmonary fibrosis. Redox Biol. 2019, 26, 101307. [Google Scholar] [CrossRef] [PubMed]
- Raffaello, A.; De Stefani, D.; Sabbadin, D.; Teardo, E.; Merli, G.; Picard, A.; Checchetto, V.; Moro, S.; Szabo, I.; Rizzuto, R. The mitochondrial calcium uniporter is a multimer that can include a dominant-negative pore-forming subunit. EMBO J. 2013, 32, 2362–2376. [Google Scholar] [CrossRef] [Green Version]
- Watano, T.; Harada, Y.; Harada, K.; Nishimura, N. Effect of Na+/Ca2+ exchange inhibitor, KB-R7943 on ouabain-induced arrhythmias in guinea-pigs. Br. J. Pharmacol. 1999, 127, 1846–1850. [Google Scholar] [CrossRef]
- DiPolo, R.; Beauge, L. Sodium/calcium exchanger: Influence of metabolic regulation on ion carrier interactions. Physiol. Rev. 2006, 86, 155–203. [Google Scholar] [CrossRef] [PubMed]
- Tedesco, S.; Adorni, M.P.; Ronda, N.; Cappellari, R.; Mioni, R.; Barbot, M.; Pinelli, S.; Plebani, M.; Bolego, C.; Scaroni, C.; et al. Activation profiles of monocyte-macrophages and HDL function in healthy women in relation to menstrual cycle and in polycystic ovary syndrome patients. Endocrine 2019. Epub Ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Toniolo, A.; Fadini, G.P.; Tedesco, S.; Cappellari, R.; Vegeto, E.; Maggi, A.; Avogaro, A.; Bolego, C.; Cignarella, A. Alternative activation of human macrophages is rescued by estrogen treatment in vitro and impaired by menopausal status. J. Clin. Endocrinol. Metab. 2015, 100, E50–E58. [Google Scholar] [CrossRef] [PubMed]
- Khananshvili, D. Sodium-calcium exchangers (NCX): Molecular hallmarks underlying the tissue-specific and systemic functions. Pflugers Arch. 2014, 466, 43–60. [Google Scholar] [CrossRef] [PubMed]
- Brustovetsky, T.; Brittain, M.K.; Sheets, P.L.; Cummins, T.R.; Pinelis, V.; Brustovetsky, N. KB-R7943, an inhibitor of the reverse Na+/Ca2+ exchanger, blocks N-methyl-D-aspartate receptor and inhibits mitochondrial complex I. Br. J. Pharmacol. 2011, 162, 255–270. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Subramanian, M.; Yurdagul, A., Jr.; Barbosa-Lorenzi, V.C.; Cai, B.; De Juan-Sanz, J.; Ryan, T.A.; Nomura, M.; Maxfield, F.R.; Tabas, I. Mitochondrial Fission Promotes the Continued Clearance of Apoptotic Cells by Macrophages. Cell 2017, 171, 331–345.e22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Repnik, U.; Knezevic, M.; Jeras, M. Simple and cost-effective isolation of monocytes from buffy coats. J. Immunol. Methods 2003, 278, 283–292. [Google Scholar] [CrossRef]
- Tedesco, S.; Bolego, C.; Toniolo, A.; Nassi, A.; Fadini, G.P.; Locati, M.; Cignarella, A. Phenotypic activation and pharmacological outcomes of spontaneously differentiated human monocyte-derived macrophages. Immunobiology 2015, 220, 545–554. [Google Scholar] [CrossRef]
- Fadini, G.P.; De Kreutzenberg, S.V.; Boscaro, E.; Albiero, M.; Cappellari, R.; Krankel, N.; Landmesser, U.; Toniolo, A.; Bolego, C.; Cignarella, A.; et al. An unbalanced monocyte polarisation in peripheral blood and bone marrow of patients with type 2 diabetes has an impact on microangiopathy. Diabetologia 2013, 56, 1856–1866. [Google Scholar] [Green Version]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef]
- Schrijvers, D.M.; Martinet, W.; De Meyer, G.R.; Andries, L.; Herman, A.G.; Kockx, M.M. Flow cytometric evaluation of a model for phagocytosis of cells undergoing apoptosis. J. Immunol. Methods 2004, 287, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Troegeler, A.; Lastrucci, C.; Duval, C.; Tanne, A.; Cougoule, C.; Maridonneau-Parini, I.; Neyrolles, O.; Lugo-Villarino, G. An efficient siRNA-mediated gene silencing in primary human monocytes, dendritic cells and macrophages. Immunol. Cell. Biol. 2014, 92, 699–708. [Google Scholar] [CrossRef] [PubMed]
- Costes, S.V.; Daelemans, D.; Cho, E.H.; Dobbin, Z.; Pavlakis, G.; Lockett, S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J. 2004, 86, 3993–4003. [Google Scholar] [CrossRef] [PubMed]
- Valente, A.J.; Maddalena, L.A.; Robb, E.L.; Moradi, F.; Stuart, J.A. A simple ImageJ macro tool for analyzing mitochondrial network morphology in mammalian cell culture. Acta Histochem. 2017, 119, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Ciubotaru, C.D.; Bastianello, S.; Beltramello, M.; Pozzan, T.; Mammano, F. Multi-Modal Imaging of Cytosolic and Mitochondrial Ca2+; Adlassnig, K.-P., Bracale, M., Eds.; ACTA Press: Anaheim, CA, USA, 2005; pp. 99–102. [Google Scholar]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tedesco, S.; Scattolini, V.; Albiero, M.; Bortolozzi, M.; Avogaro, A.; Cignarella, A.; Fadini, G.P. Mitochondrial Calcium Uptake Is Instrumental to Alternative Macrophage Polarization and Phagocytic Activity. Int. J. Mol. Sci. 2019, 20, 4966. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20194966
Tedesco S, Scattolini V, Albiero M, Bortolozzi M, Avogaro A, Cignarella A, Fadini GP. Mitochondrial Calcium Uptake Is Instrumental to Alternative Macrophage Polarization and Phagocytic Activity. International Journal of Molecular Sciences. 2019; 20(19):4966. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20194966
Chicago/Turabian StyleTedesco, Serena, Valentina Scattolini, Mattia Albiero, Mario Bortolozzi, Angelo Avogaro, Andrea Cignarella, and Gian Paolo Fadini. 2019. "Mitochondrial Calcium Uptake Is Instrumental to Alternative Macrophage Polarization and Phagocytic Activity" International Journal of Molecular Sciences 20, no. 19: 4966. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20194966