Glycyrrhizin Acid and Glycyrrhetinic Acid Modified Polyethyleneimine for Targeted DNA Delivery to Hepatocellular Carcinoma
Abstract
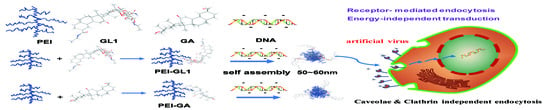
:1. Introduction
2. Results and Discussion
2.1. Synthesis and Characterization of GA or GL Functionalized PEIs
2.2. Biophysical Characterization of GA or GL Modified PEIs
2.3. Cytotoxicity and Gene Transfection in Vitro
2.4. Effect of free GA on the Transfection Efficiency
2.5. Endocytic Pathway and Intracellular Trafficking
2.6. In Vivo Gene Transfection
3. Materials and Methods
3.1. Materials and Cell Culture
3.2. Preparation and Characterization of PEI-GA and PEI-GL Polyplexes
3.3. Gel Retardation Assay
3.4. Cytotoxicity of PEI-GA and PEI-GL Conjugates
3.5. Evaluation of Transfection Efficiency in Vitro
3.6. Free GA Competition Assay
3.7. Endocytosis Inhibitor Effects on Cellular Uptake and Transfection
3.8. Observation of Intracellular Trafficking by Confocal Imaging
3.9. Transfection in Vivo
3.10. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Cai, Y.; Xu, Y.; Chan, H.F.; Fang, X.; He, C.; Chen, M. Glycyrrhetinic Acid Mediated Drug Delivery Carriers for Hepatocellular Carcinoma Therapy. Mol. Pharm. 2016, 13, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Reghupaty, S.C.; Sarkar, D. Current Status of Gene Therapy in Hepatocellular Carcinoma. Cancers (Basel) 2019, 11, 1265. [Google Scholar] [CrossRef] [PubMed]
- Rettig, G.R.; Behlke, M.A. Progress Toward In Vivo Use of siRNAs-II. Mol. Ther. 2012, 20, 483–512. [Google Scholar] [CrossRef] [PubMed]
- Kaneshiro, T.L.; Lu, Z.-R. Targeted intracellular codelivery of chemotherapeutics and nucleic acid with a well-defined dendrimer-based nanoglobular carrier. Biomaterials 2009, 30, 5660–5666. [Google Scholar] [CrossRef] [PubMed]
- Rezaee, M.; Oskuee, R.K.; Nassirli, H.; Malaekeh-Nikouei, B. Progress in the development of lipopolyplexes as efficient non-viral gene delivery systems. J. Control. Release 2016, 236, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Merdan, T.; Kopeček, J.; Kissel, T. Prospects for cationic polymers in gene and oligonucleotide therapy against cancer. Adv. Drug Deliv. Rev. 2002, 54, 715–758. [Google Scholar] [CrossRef]
- Morille, M.; Passirani, C.; Vonarbourg, A.; Clavreul, A.; Benoit, J.-P. Progress in developing cationic vectors for non-viral systemic gene therapy against cancer. Biomaterials 2008, 29, 3477–3496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, H.; Kanasty, R.L.; Eltoukhy, A.A.; Vegas, A.J.; Dorkin, J.R.; Anderson, D.G. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014, 15, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Godbey, W.T.; Wu, K.K.; Mikos, A.G. Poly(ethylenimine) and its role in gene delivery. J. Control. Release 1999, 60, 149–160. [Google Scholar] [CrossRef]
- Piest, M.; Engbersen, J.F. Effects of charge density and hydrophobicity of poly(amido amine)s for non-viral gene delivery. J. Control. Release 2010, 148, 83–90. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Wang, Z.; Wang, F.; He, J.; Yang, X.; Xie, W.; Liu, Y.; Zhang, Y. A thermosensitive gel based on w1/o/w2 multiple microemulsions for the vaginal delivery of small nucleic acid. Drug Deliv. 2019, 26, 168–178. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yu, M.; Wang, J.; Tang, R.; Yan, G.; Yao, W. Low Molecular Weight PEI-Based Vectors via Acid-Labile Ortho Ester Linkage for Improved Gene Delivery. Macromol. Biosci. 2016, 16, 1175–1187. [Google Scholar] [CrossRef] [PubMed]
- Gomez, J.P.; Tresset, G.; Pichon, C.; Midoux, P. Improved histidinylated lPEI polyplexes for skeletal muscle cells transfection. Int. J. Pharm. 2019, 559, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Tian, H.; Dong, X.; Guo, Z.; Jiao, Z.; Li, F.; Kano, A.; Maruyama, A.; Chen, X. Effective tumor treatment by VEGF siRNA complexed with hydrophobic poly(amino acid)-modified polyethylenimine. Macromol. Biosci. 2013, 13, 1438–1446. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Liu, Y.; Gao, H.; Fei, H.; Yu, W.; Hu, T.; Zheng, Y.; Bia, X.; Lin, C. N-Acetyl-l-leucine-polyethylenimine-mediated miR-34a delivery improves osteogenesis and bone formation in vitro and in vivo. RSC Adv. 2018, 8, 8080–8088. [Google Scholar] [CrossRef]
- Teo, P.Y.; Yang, C.; Hedrick, J.L.; Engler, A.C.; Coady, D.J.; Ghaem-Maghami, S.; George, A.J.; Yang, Y.Y. Hydrophobic modification of low molecular weight polyethylenimine for improved gene transfection. Biomaterials 2013, 34, 7971–7979. [Google Scholar] [CrossRef] [PubMed]
- Chae, S.Y.; Kim, H.J.; Lee, M.S.; Jang, Y.L.; Lee, Y.; Lee, S.H.; Lee, K.; Kim, S.H.; Kim, H.T.; Chi, S.-C.; et al. Energy-independent intracellular gene delivery mediated by polymeric biomimetics of cell-penetrating peptides. Macromol. Biosci. 2011, 11, 1169–1174. [Google Scholar] [CrossRef]
- Thapa, B.; Plianwong, S.; Bahadur, K.R.; Rutherford, B.; Uludağ, H. Small hydrophobe substitution on polyethylenimine for plasmid DNA delivery: Optimal substitution is critical for effective delivery. Acta Biomater. 2016, 33, 213–224. [Google Scholar] [CrossRef]
- Yameen, B.; Choi, W.I.; Vilos, C.; Swami, A.; Shi, J.; Farokhzad, O.C. Insight into nanoparticle cellular uptake and intracellular targeting. J. Control. Release 2014, 190, 485–499. [Google Scholar] [CrossRef] [Green Version]
- Ulbrich, K.; Holá, K.; Šubr, V.; Bakandritsos, A.; Tuček, J.; Zboril, R. Targeted Drug Delivery with Polymers and Magnetic Nanoparticles: Covalent and Noncovalent Approaches, Release Control, and Clinical Studies. Chem. Rev. 2016, 116, 5338–5431. [Google Scholar] [CrossRef]
- Hu, Y.; Xu, B.; Ji, Q.; Shou, D.; Sun, X.; Xu, J.; Gao, J.; Liang, W. A mannosylated cell-penetrating peptide-graft-polyethylenimine as a gene delivery vector. Biomaterials 2014, 35, 4236–4246. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Su, J.; Cai, W.; Lu, P.; Yuan, L.; Jin, T.; Chen, S.; Sheng, J. Hepatocyte-targeting gene transfer mediated by galactosylated poly(ethylene glycol)-graft-polyethylenimine derivative. Drug Des. Dev. Ther. 2013, 7, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Kos, P.; Lächelt, U.; He, D.; Nie, Y.; Gu, Z.; Wagner, E. Dual-targeted polyplexes based on sequence-defined peptide-PEG-oligoamino amides. J. Pharm. Sci. 2015, 104, 464–475. [Google Scholar] [CrossRef] [PubMed]
- Hibasami, H.; Iwase, H.; Yoshioka, K.; Takahashi, H. Glycyrrhetic acid (a metabolic substance and aglycon of glycyrrhizin) induces apoptosis in human hepatoma, promyelotic leukemia and stomach cancer cells. Int. J. Mol. Med. 2006, 17, 215–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Negishi, M.; Irie, A.; Nagata, N.; Ichikawa, A. Specific binding of glycyrrhetinic acid to the rat liver membrane. Biochim. Biophys. Acta (BBA) Biomembr. 1991, 1066, 77–82. [Google Scholar] [CrossRef]
- Ishida, S.; Sakiya, Y.; Ichikawa, T.; Taira, Z. Uptake of glycyrrhizin by isolated rat hepatocytes. Biol. Pharm. Bull. 1993, 16, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Tang, C.; Yin, C. Glycyrrhizin-modified O-carboxymethyl chitosan nanoparticles as drug vehicles targeting hepatocellular carcinoma. Biomaterials 2012, 33, 7594–7604. [Google Scholar] [CrossRef]
- Lin, A.; Liu, Y.; Huang, Y.; Sun, J.; Wu, Z.; Zhang, X.; Ping, Q. Glycyrrhizin surface-modified chitosan nanoparticles for hepatocyte-targeted delivery. Int. J. Pharm. 2008, 359, 247–253. [Google Scholar] [CrossRef]
- Chopdey, P.K.; Tekade, R.K.; Mehra, N.K.; Mody, N.; Jain, N.K. Glycyrrhizin Conjugated Dendrimer and Multi-Walled Carbon Nanotubes for Liver Specific Delivery of Doxorubicin. J. Nanosci. Nanotechnol. 2015, 15, 1088–1100. [Google Scholar] [CrossRef]
- Mishra, D.; Jain, N.; Rajoriya, V.; Jain, A.K. Glycyrrhizin conjugated chitosan nanoparticles for hepatocyte-targeted delivery of lamivudine. J. Pharm. Pharmacol. 2014, 66, 1082–1093. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, W.; Liu, T.; Wu, Y.; Guo, H.; Wang, P.; Tian, Q.; Wang, Y.; Yuan, Z. Doxorubicin-loaded glycyrrhetinic acid-modified alginate nanoparticles for liver tumor chemotherapy. Biomaterials 2012, 33, 2187–2196. [Google Scholar] [CrossRef] [PubMed]
- Meneksedag-Erol, D.; Kc, R.B.; Tang, T.; Uludağ, H. A Delicate Balance When Substituting a Small Hydrophobe onto Low Molecular Weight Polyethylenimine to Improve Its Nucleic Acid Delivery Efficiency. ACS Appl. Mater. Interfaces 2015, 7, 24822–24832. [Google Scholar] [CrossRef] [PubMed]
- Cong, Y.; Shi, B.; Lu, Y.; Wen, S.; Chung, R.; Jin, D. One-step Conjugation of Glycyrrhetinic Acid to Cationic Polymers for High-performance Gene Delivery to Cultured Liver Cell. Sci. Rep. 2016, 6, 21891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, S.P.; Gabrielson, N.P.; Wong, C.-H.; Chow, H.-F.; Pack, D.W.; Posocco, P.; Fermeglia, M.; Pricl, S.; Smith, D.K. Hydrophobically modified dendrons: developing structure-activity relationships for DNA binding and gene transfection. Mol. Pharm. 2011, 8, 416–429. [Google Scholar] [CrossRef] [PubMed]
- Dahlman, J.E.; Barnes, C.; Khan, O.F.; Thiriot, A.; Jhunjunwala, S.; Shaw, T.E.; Xing, Y.; Sager, H.B.; Sahay, G.; Speciner, L.; et al. In vivo endothelial siRNA delivery using polymeric nanoparticles with low molecular weight. Nat. Nanotechnol. 2014, 9, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Yuan, Z.; Xu, Y.; Zhao, X.; Fang, Z.; Yuan, W.-E. A Low-Molecular-Weight Polyethylenimine/pDNA-VEGF Polyplex System Constructed in a One-Pot Manner for Hindlimb Ischemia Therapy. Pharmaceutics 2019, 11, 171. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, W.; Huang, Y.; Gao, F.; Sha, X.; Lou, K.-Y.; Fang, X. The therapeutic effect of methotrexate-conjugated Pluronic-based polymeric micelles on the folate receptor-rich tumors treatment. Int. J. Nanomed. 2015, 10, 4043–4057. [Google Scholar] [CrossRef]
- Qiu, N.; Liu, X.; Zhong, Y.; Zhou, Z.; Piao, Y.; Miao, L.; Zhang, Q.; Tang, J.; Huang, L.; Shen, Y. Esterase-Activated Charge-Reversal Polymer for Fibroblast-Exempt Cancer Gene Therapy. Adv. Mater. 2016, 28, 10613–10622. [Google Scholar] [CrossRef]
- Guo, Z.; Tian, H.; Lin, L.; Chen, J.; He, C.; Tang, Z.; Chen, X. Hydrophobic polyalanine modified hyperbranched polyethylenimine as high efficient pDNA and siRNA carrier. Macromol. Biosci. 2014, 14, 1406–1414. [Google Scholar] [CrossRef]
- Feng, R.; Deng, P.; Song, Z.; Chu, W.; Zhu, W.; Teng, F.; Zhou, F. Glycyrrhetinic acid-modified PEG-PCL copolymeric micelles for the delivery of curcumin. React. Funct. Polymers 2017, 111, 30–37. [Google Scholar] [CrossRef]
- Zhang, L.; Yao, J.; Zhou, J.; Wang, T.; Zhang, Q. Glycyrrhetinic acid-graft-hyaluronic acid conjugate as a carrier for synergistic targeted delivery of antitumor drugs. Int. J. Pharm. 2013, 441, 654–664. [Google Scholar] [CrossRef] [PubMed]
- Guo, G.; Zhou, L.; Chen, Z.; Chi, W.; Yang, X.; Wang, W.; Zhang, B. Alkane-modified low-molecular-weight polyethylenimine with enhanced gene silencing for siRNA delivery. Int. J. Pharm. 2013, 450, 44–52. [Google Scholar] [CrossRef] [PubMed]
- Ganesh, S.; Iyer, A.K.; Morrissey, D.V.; Amiji, M.M. Hyaluronic acid based self-assembling nanosystems for CD44 target mediated siRNA delivery to solid tumors. Biomaterials 2013, 34, 3489–3502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, Y.; Zhao, M.; Chen, Y.; Hua, L.; Xu, T.; Wang, C.; Li, Y.; Zhu, B. Folate-targeted selenium nanoparticles deliver therapeutic siRNA to improve hepatocellular carcinoma therapy. RSC Adv. 2018, 8, 25932–25940. [Google Scholar] [CrossRef] [Green Version]
- Zou, Y.; Song, Y.; Yang, W.; Meng, F.; Liu, H.; Zhong, Z. Galactose-installed photo-crosslinked pH-sensitive degradable micelles for active targeting chemotherapy of hepatocellular carcinoma in mice. J. Control. Release 2014, 193, 154–161. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, M.; Yung, B.; Li, H.; Zhou, C.; Lee, L.J.; Lee, R.J. Lactosylated liposomes for targeted delivery of doxorubicin to hepatocellular carcinoma. Int. J. Nanomed. 2012, 7, 5465–5474. [Google Scholar] [Green Version]
- Li, Y.; Lee, R.J.; Yu, K.; Bi, Y.; Qi, Y.; Sun, Y.; Li, Y.; Xie, J.; Teng, L. Delivery of siRNA Using Lipid Nanoparticles Modified with Cell Penetrating Peptide. ACS Appl. Mater. Interfaces 2016, 8, 26613–26621. [Google Scholar] [CrossRef]
- Kurosaki, T.; Kawanabe, S.; Kodama, Y.; Fumoto, S.; Nishida, K.; Nakagawa, H.; Higuchi, N.; Nakamura, T.; Kitahara, T.; Sasaki, H. Hepatic gene delivery system electrostatically assembled with glycyrrhizin. Mol. Pharm. 2014, 11, 1369–1377. [Google Scholar] [CrossRef]
- Gabrielson, N.P.; Pack, D.W. Efficient polyethylenimine-mediated gene delivery proceeds via a caveolar pathway in HeLa cells. J. Control. Release 2009, 136, 54–61. [Google Scholar] [CrossRef] [Green Version]
- Shen, W.; Van Dongen, M.A.; Han, Y.; Yu, M.; Li, Y.; Liu, G.; Holl, M.M.B.; Qi, R. The role of caveolin-1 and syndecan-4 in the internalization of PEGylated PAMAM dendrimer polyplexes into myoblast and hepatic cells. Eur. J. Pharm. Biopharm. 2014, 88, 658–663. [Google Scholar] [CrossRef] [Green Version]
- Sahay, G.; Alakhova, D.Y.; Kabanov, A.V. Endocytosis of nanomedicines. J. Control. Release 2010, 145, 182–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varkouhi, A.K.; Scholte, M.; Storm, G.; Haisma, H.J. Endosomal escape pathways for delivery of biologicals. J. Control. Release 2011, 151, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Damm, E.M.; Pelkmans, L.; Kartenbeck, J.; Mezzacasa, A.; Kurzchalia, T.; Helenius, A. Clathrin- and caveolin-1–independent endocytosis. J. Cell Biol. 2005, 168, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Duncan, R.; Richardson, S.C.W. Endocytosis and intracellular trafficking as gateways for nanomedicine delivery: Opportunities and challenges. Mol. Pharm. 2012, 9, 2380–2402. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Chen, S.; Han, J.; Zhang, Z. Mannosylated biodegradable polyethyleneimine for targeted DNA delivery to dendritic cells. Int. J. Nanomed. 2012, 7, 2929–2942. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Xiang, J.; Zhu, D.; Jiang, L.; Zhou, Z.; Tang, J.; Liu, X.; Huang, Y.; Shen, Y. Fusogenic Reactive Oxygen Species Triggered Charge-Reversal Vector for Effective Gene Delivery. Adv. Mater. 2016, 28, 1743–1752. [Google Scholar] [CrossRef]
- Wang, J.; Meng, F.; Kim, B.-K.; Ke, X.; Yeo, Y. In-vitro and in-vivo difference in gene delivery by lithocholic acid-polyethyleneimine conjugate. Biomaterials 2019, 217, 119296. [Google Scholar] [CrossRef]
- Budker, V.G.; Monahan, S.D.; Subbotin, V.M. Loco-regional cancer drug therapy: present approaches and rapidly reversible hydrophobization (RRH) of therapeutic agents as the future direction. Drug Discov. Today 2014, 19, 1855–1870. [Google Scholar] [CrossRef] [Green Version]
- Odendahl, K.; Solass, W.; Demtröder, C.; Giger-Pabst, U.; Zieren, J.; Tempfer, C.; Reymond, M. Quality of life of patients with end-stage peritoneal metastasis treated with Pressurized IntraPeritoneal Aerosol Chemotherapy (PIPAC). Eur. J. Surg. Oncol. 2015, 41, 1379–1385. [Google Scholar] [CrossRef] [Green Version]
- Hazekawa, M.; Nishinakagawa, T.; Kawakubo-Yasukochi, T.; Nakashima, M. Glypican-3 gene silencing for ovarian cancer using siRNA-PLGA hybrid micelles in a murine peritoneal dissemination model. J. Pharmacol. Sci. 2019, 139, 231–239. [Google Scholar] [CrossRef]
- Qiu, N.; Gao, J.; Liu, Q.; Wang, J.; Shen, Y. Enzyme-Responsive Charge-Reversal Polymer-Mediated Effective Gene Therapy for Intraperitoneal Tumors. Biomacromolecules 2018, 19, 2308–2319. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Kang, X.; Yuan, B.; Wang, H.; Zhang, T.; Shi, M.; Zheng, Z.; Zhang, Y.; Peng, C.; Fan, X.; et al. Nano-Structural Effects on Gene Transfection: Large, Botryoid-Shaped Nanoparticles Enhance DNA Delivery via Macropinocytosis and Effective Dissociation. Theranostics 2019, 9, 1580–1598. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, P.; Yuan, S.; Han, S.; Mangrio, F.A.; Zhu, Z.; Lei, F.; Ming, Z.; Cheng, L.; Liu, Z.; Si, T.; et al. Core-shell microencapsulation of curcumin in PLGA microparticles: Programmed for application in ovarian cancer therapy. Artif. Cells Nanomed. Biotechnol. 2018, 46, S481–S491. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Zhong, Y.; Meng, F.; Peng, R.; Zhong, Z. Lipoic acid modified low molecular weight polyethylenimine mediates nontoxic and highly potent in vitro gene transfection. Mol. Pharm. 2011, 8, 2434–2443. [Google Scholar] [CrossRef] [PubMed]
- Hwang, M.E.; Keswani, R.K.; Pack, D.W. Dependence of PEI and PAMAM Gene Delivery on Clathrin- and Caveolin-Dependent Trafficking Pathways. Pharm. Res. 2015, 32, 2051–2059. [Google Scholar] [CrossRef] [PubMed]










| Item | Feed Ratio | Substitution Amount | Yield | |
|---|---|---|---|---|
| (by OD) | (by 1HNMR) | |||
| PEI:GA | 1:1 | 0.75 | 0.69 | 67.8% |
| 1:2 | 1.10 | 1.16 | 54.1% | |
| 1:3 | 1.71 | 1.89 | 45.8% | |
| PEI:GL1 | 1:1 | 0.62 | 0.66 | 51.6% |
| PEI:GL2 | 1:1 | 0.65 | 0.67 | 52.8% |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, M.; Gao, Y.; Zhan, M.; Qiu, N.; Piao, Y.; Zhou, Z.; Shen, Y. Glycyrrhizin Acid and Glycyrrhetinic Acid Modified Polyethyleneimine for Targeted DNA Delivery to Hepatocellular Carcinoma. Int. J. Mol. Sci. 2019, 20, 5074. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205074
Cao M, Gao Y, Zhan M, Qiu N, Piao Y, Zhou Z, Shen Y. Glycyrrhizin Acid and Glycyrrhetinic Acid Modified Polyethyleneimine for Targeted DNA Delivery to Hepatocellular Carcinoma. International Journal of Molecular Sciences. 2019; 20(20):5074. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205074
Chicago/Turabian StyleCao, Mingzhuo, Yong Gao, Mengling Zhan, Nasha Qiu, Ying Piao, Zhuxian Zhou, and Youqing Shen. 2019. "Glycyrrhizin Acid and Glycyrrhetinic Acid Modified Polyethyleneimine for Targeted DNA Delivery to Hepatocellular Carcinoma" International Journal of Molecular Sciences 20, no. 20: 5074. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205074