Gene Expression Profiling of the Extracellular Matrix Signature in Macrophages of Different Activation Status: Relevance for Skin Wound Healing
Abstract
:1. Introduction
2. Results
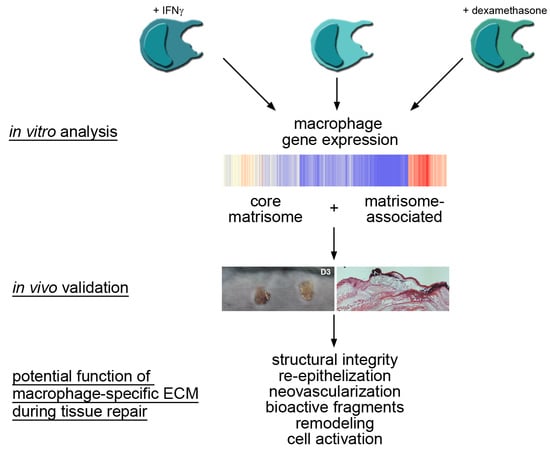
2.1. Core Matrisome and Matrisome-Associated Genes Are Expressed in Human Macrophages
2.2. A Unique Panel of Core Matrisome and Matrisome-Associated Genes Is Regulated in Interferon Gamma (IFNγ)- and Dexamethasone-Primed Macrophages
3. Discussion
3.1. Macrophages Could Provide Structural Integrity by Expressing Core Matrisome Genes
3.2. Distinct Core Matrisome Genes That Are Induced in Dexamethasone-Stimulated Macrophages Contribute to Re-Epithelization and Neovascularization
3.3. Macrophages Express Genes That Are Involved in the Release of Bioactive Fragments from the Extracellular Matrix (ECM)
4. Materials and Methods
4.1. Bioinformatic Analysis
4.2. Wound-Healing Experiments
4.3. Antibody Generation
4.4. Immunofluorescence Analysis
4.5. Immunoblot Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ADAM | A disintegrin and metalloproteinase |
| ADAMTS | ADAM with thrombospondin motifs |
| ADAMTSL | ADAMTS like |
| AGRN | Agrin |
| A2M | Alpha-2-macroglobulin |
| ANXA | Annexin |
| BMP | Bone morphogenetic protein |
| BRAL | Brain link protein |
| CILP | Cartilage intermediate layer protein |
| CTS | Cathepsin |
| CXCL | Chemokine (C-X-C motif) ligand |
| CCL | Chemokine ligand |
| CHADL | Chondroadherin-like |
| CD | Cluster of differentiation |
| COL | Collagen |
| C1Q | Complement component 1q |
| CLEC | C-type lectin domain family |
| CST | Cystatin |
| Dexa | Dexamethasone |
| EMILIN | Elastin microfibril interfacer |
| EMT | Epithelial-to-mesenchymal transition |
| ECM | Extracellular matrix |
| FAM | Family with sequence similarity |
| FGL | Fibrinogen-like |
| FGF | Fibroblast growth factor |
| FBLN | Fibulin |
| FCN | Ficolin |
| FC | Fold change |
| FDR | False discovery rate |
| GLDN | Gliomedin |
| GREM | Gremlin |
| GDF | Growth/differentiation factor |
| HTR | High-temperature requirement |
| HAPLN | Hyaluronan and proteoglycan link protein |
| HA | Hyaluronan |
| HYAL | Hyaluronidase |
| INHB | Inhibin subunit beta |
| IGF | Insulin-like growth factor |
| ITGA | Integrin alpha subunit |
| ITGB | Integrin beta subunit |
| IFNγ | Interferon gamma |
| IL | Interleukin |
| LAMA | Laminin alpha chain |
| LAMB | Laminin beta chain |
| LAMC | Laminin gamma chain |
| LGALS | Lectin, galactoside-binding, soluble |
| LPS | Lipopolysaccharides |
| LH | Lysyl hydroxylase |
| MCSF | Macrophage colony-stimulating factor |
| MMP | Matrix metalloproteinase |
| MFGE | Milk fat globule epidermal growth fator |
| MDM | Monocyte-derived macrophage |
| NTN | Netrin |
| ON | Osteonectin |
| PLAU | Plasminogen activator, urokinase |
| PDGF | Platelet-derived growth factor |
| PLXN | Plexin |
| PLXDC | Plexin domain containing |
| PCOLCE | Procollagen C-endopeptidase enhancer |
| PPBP | Pro-platelet basic protein |
| SPP1 | Secreted phosphoprotein 1 |
| SPARC | Secreted protein acidic and rich in cysteines |
| SEMA | Semaphorin |
| SPOCK | SPARC/osteonectin, Cwcv, and Kazal-like domains proteoglycan |
| SRPX | Sushi repeat-containing protein |
| THBS | Thrombospondin |
| TIMP | Tissue inhibitors of metalloproteinases |
| TLR | Toll-like receptor |
| TGFβ | Transforming growth factor beta |
| TGFBI | Transforming growth factor beta induced |
| TINAGL1 | Tubulointerstitial nephritis antigen-like |
| TSG | Tumor necrosis factor stimulated gene |
| TNFAIP | Tumor necrosis factor alpha induced protein |
| TAM | Tumor-associated macrophage |
| UNC | Uncoordinated locomotion |
| VEGF | Vascular-endothelial growth factor |
| VCAN | Versican |
References
- Grskovic, I.; Kutsch, A.; Frie, C.; Groma, G.; Stermann, J.; Schlötzer-Schrehardt, U.; Niehoff, A.; Moss, S.E.; Rosenbaum, S.; Pöschl, E.; et al. Depletion of annexin A5, annexin A6, and collagen X causes no gross changes in matrix vesicle-mediated mineralization, but lack of collagen X affects hematopoiesis and the Th1/Th2 response. J. Bone Miner. Res. 2012, 27, 2399–2412. [Google Scholar]
- Probst, K.; Stermann, J.; von Bomhard, I.; Etich, J.; Pitzler, L.; Niehoff, A.; Bluhm, B.; Xu, H.C.; Lang, P.A.; Chmielewski, M.; et al. Depletion of Collagen IX Alpha1 Impairs Myeloid Cell Function. Stem Cells 2018, 36, 1752–1763. [Google Scholar] [Green Version]
- Sweeney, E.; Campbell, M.; Watkins, K.; Hunter, C.A.; Jacenko, O. Altered endochondral ossification in collagen X mouse models leads to impaired immune responses. Dev. Dyn. 2008, 237, 2693–2704. [Google Scholar] [Green Version]
- Smigiel, K.S.; Parks, W.C. Macrophages, Wound Healing, and Fibrosis: Recent Insights. Curr. Rheumatol. Rep. 2018, 20, 17. [Google Scholar]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage Activation and Polarization: Nomenclature and Experimental Guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [Green Version]
- Wight, T.N.; Kang, I.; Merrilees, M.J. Versican and the control of inflammation. Matrix Biol. 2014, 35, 152–161. [Google Scholar]
- Sicari, B.M.; Dziki, J.L.; Siu, B.F.; Medberry, C.J.; Dearth, C.L.; Badylak, S.F. The promotion of a constructive macrophage phenotype by solubilized extracellular matrix. Biomaterials 2014, 35, 8605–8612. [Google Scholar]
- Gaudet, A.D.; Popovich, P.G. Extracellular matrix regulation of inflammation in the healthy and injured spinal cord. Exp. Neurol. 2014, 258, 24–34. [Google Scholar] [Green Version]
- Kim, S.Y.; Nair, M.G. Macrophages in wound healing: Activation and plasticity. Immunol. Cell Biol. 2019, 97, 258–267. [Google Scholar]
- Navegantes, K.C.; de Souza Gomes, R.; Pereira, P.A.T.; Czaikoski, P.G.; Azevedo, C.H.M.; Monteiro, M.C. Immune modulation of some autoimmune diseases: The critical role of macrophages and neutrophils in the innate and adaptive immunity. J. Transl. Med. 2017, 15, 36. [Google Scholar]
- Barron, L.; Wynn, T.A. Fibrosis is regulated by Th2 and Th17 responses and by dynamic interactions between fibroblasts and macrophages. Am. J. Physiol. Gastrointest. Liver Physiol. 2011, 300, G723–G728. [Google Scholar] [Green Version]
- Shook, B.A.; Wasko, R.R.; Rivera-Gonzalez, G.C.; Salazar-Gatzimas, E.; López-Giráldez, F.; Dash, B.C.; Muñoz-Rojas, A.R.; Aultman, K.D.; Zwick, R.K.; Lei, V.; et al. Myofibroblast proliferation and heterogeneity are supported by macrophages during skin repair. Science 2018, 362, eaar2971. [Google Scholar]
- Knipper, J.A.; Willenborg, S.; Brinckmann, J.; Bloch, W.; Maaß, T.; Wagener, R.; Krieg, T.; Sutherland, T.; Munitz, A.; Rothenberg, M.E.; et al. Interleukin-4 Receptor α Signaling in Myeloid Cells Controls Collagen Fibril Assembly in Skin Repair. Immunity 2015, 43, 803. [Google Scholar]
- Bergmeier, V.; Etich, J.; Pitzler, L.; Frie, C.; Koch, M.; Fischer, M.; Rappl, G.; Abken, H.; Tomasek, J.J.; Brachvogel, B. Identification of a myofibroblast-specific expression signature in skin wounds. Matrix Biol. 2018, 65, 59–74. [Google Scholar]
- Piccinini, A.M.; Zuliani-Alvarez, L.; Lim, J.M.P.; Midwood, K.S. Distinct microenvironmental cues stimulate divergent TLR4-mediated signaling pathways in macrophages. Sci. Signal. 2016, 9, ra86. [Google Scholar]
- Schnoor, M.; Cullen, P.; Lorkowski, J.; Stolle, K.; Robenek, H.; Troyer, D.; Rauterberg, J.; Lorkowski, S. Production of type VI collagen by human macrophages: A new dimension in macrophage functional heterogeneity. J. Immunol. 2008, 180, 5707–5719. [Google Scholar]
- Weitkamp, B.; Cullen, P.; Plenz, G.; Robenek, H.; Rauterberg, J. Human macrophages synthesize type VIII collagen in vitro and in the atherosclerotic plaque. FASEB J. 1999, 13, 1445–1457. [Google Scholar]
- Chang, M.Y.; Chan, C.K.; Braun, K.R.; Green, P.S.; O’Brien, K.D.; Chait, A.; Day, A.J.; Wight, T.N. Monocyte-to-macrophage differentiation: Synthesis and secretion of a complex extracellular matrix. J. Biol. Chem. 2012, 287, 14122–14135. [Google Scholar]
- Steiger, J.; Stephan, A.; Inkeles, M.S.; Realegeno, S.; Bruns, H.; Kröll, P.; de Castro Kroner, J.; Sommer, A.; Batinica, M.; Pitzler, L.; et al. Imatinib Triggers Phagolysosome Acidification and Antimicrobial Activity against Mycobacterium bovis Bacille Calmette-Guérin in Glucocorticoid-Treated Human Macrophages. J. Immunol. 2016, 197, 222–232. [Google Scholar]
- Naba, A.; Clauser, K.R.; Ding, H.; Whittaker, C.A.; Carr, S.A.; Hynes, R.O. The extracellular matrix: Tools and insights for the “omics” era. Matrix Biol. 2016, 49, 10–24. [Google Scholar]
- Hynes, R.O.; Naba, A. Overview of the matrisome--an inventory of extracellular matrix constituents and functions. Cold Spring Harb. Perspect. Biol. 2012, 4, a004903. [Google Scholar]
- Tajiri, Y.; Igarashi, T.; Li, D.; Mukai, K.; Suematsu, M.; Fukui, E.; Yoshizawa, M.; Matsumoto, H. Tubulointerstitial Nephritis Antigen-Like 1 Is Expressed in the Uterus and Binds with Integrins in Decidualized Endometrium During Postimplantation in Mice1. Biol. Reprod. 2009, 82, 263–270. [Google Scholar]
- Kim, J.E.; Kim, S.J.; Lee, B.H.; Park, R.W.; Kim, K.S.; Kim, I.S. Identification of motifs for cell adhesion within the repeated domains of transforming growth factor-β-induced gene, βig-h3. J. Biol. Chem. 2000, 275, 30907–30915. [Google Scholar]
- Erikson, D.W.; Burghardt, R.C.; Bayless, K.J.; Johnson, G.A. Secreted Phosphoprotein 1 (SPP1, Osteopontin) Binds to Integrin Alphavbeta6 on Porcine Trophectoderm Cells and Integrin Alphavbeta3 on Uterine Luminal Epithelial Cells, and Promotes Trophectoderm Cell Adhesion and Migration1. Biol. Reprod. 2009, 81, 814–825. [Google Scholar] [Green Version]
- Verdone, G.; Doliana, R.; Corazza, A.; Colebrooke, S.A.; Spessotto, P.; Bot, S.; Bucciotti, F.; Capuano, A.; Silvestri, A.; Viglino, P.; et al. The solution structure of EMILIN1 globular C1q domain reveals a disordered insertion necessary for interaction with the α4β1 integrin. J. Biol. Chem. 2008, 283, 18947–18956. [Google Scholar]
- Sangaletti, S.; Di Carlo, E.; Gariboldi, S.; Miotti, S.; Cappetti, B.; Parenza, M.; Rumio, C.; Brekken, R.A.; Chiodoni, C.; Colombo, M.P. Macrophage-Derived SPARC Bridges Tumor Cell-Extracellular Matrix Interactions toward Metastasis. Cancer Res. 2008, 68, 9050–9059. [Google Scholar] [Green Version]
- Nishiuchi, R.; Takagi, J.; Hayashi, M.; Ido, H.; Yagi, Y.; Sanzen, N.; Tsuji, T.; Yamada, M.; Sekiguchi, K. Ligand-binding specificities of laminin-binding integrins: A comprehensive survey of laminin–integrin interactions using recombinant α3β1, α6β1, α7β1 and α6β4 integrins. Matrix Biol. 2006, 25, 189–197. [Google Scholar]
- Janssen, B.J.C.; Robinson, R.A.; Pérez-Brangulí, F.; Bell, C.H.; Mitchell, K.J.; Siebold, C.; Jones, E.Y. Structural basis of semaphorin–plexin signalling. Nature 2010, 467, 1118–1122. [Google Scholar]
- Nogi, T.; Yasui, N.; Mihara, E.; Matsunaga, Y.; Noda, M.; Yamashita, N.; Toyofuku, T.; Uchiyama, S.; Goshima, Y.; Kumanogoh, A.; et al. Structural basis for semaphorin signalling through the plexin receptor. Nature 2010, 467, 1123–1127. [Google Scholar]
- Kim, H.; Chung, H.; Kim, J.; Choi, D.-H.; Shin, Y.; Kang, Y.G.; Kim, B.-M.; Seo, S.-U.; Chung, S.; Seok, S.H. Macrophages-Triggered Sequential Remodeling of Endothelium-Interstitial Matrix to Form Pre-Metastatic Niche in Microfluidic Tumor Microenvironment. Adv. Sci. (Weinh) 2019, 6, 1900195. [Google Scholar]
- Albeiroti, S.; Ayasoufi, K.; Hill, D.R.; Shen, B.; de la Motte, C.A. Platelet hyaluronidase-2: An enzyme that translocates to the surface upon activation to function in extracellular matrix degradation. Blood 2015, 125, 1460–1469. [Google Scholar]
- Kreft, S.; Klatt, A.R.; Straßburger, J.; Pöschl, E.; Flower, R.J.; Eming, S.; Reutelingsperger, C.; Brisson, A.; Brachvogel, B. Skin wound repair is not altered in the absence of endogenous AnxA1 or AnxA5, but pharmacological concentrations of AnxA4 and AnxA5 inhibit wound hemostasis. Cells Tissues Organs 2016, 201, 287–298. [Google Scholar]
- Shechter, R.; Schwartz, M. CNS sterile injury: Just another wound healing? Trends Mol. Med. 2013, 19, 135–143. [Google Scholar]
- Kadler, K.E.; Hill, A.; Canty-Laird, E.G. Collagen fibrillogenesis: Fibronectin, integrins, and minor collagens as organizers and nucleators. Curr. Opin. Cell Biol. 2008, 20, 495–501. [Google Scholar]
- Yoshioka, H.; Iyama, K.; Inoguchi, K.; Khaleduzzaman, M.; Ninomiya, Y.; Ramirez, F. Developmental pattern of expression of the mouse alpha 1 (XI) collagen gene (Col11a1). Dev. Dyn. 1995, 204, 41–47. [Google Scholar]
- Smith, S.M.; Birk, D.E. Focus on molecules: Collagens V and XI. Exp. Eye Res. 2012, 98, 105–106. [Google Scholar]
- Ingman, W.V.; Wyckoff, J.; Gouon-Evans, V.; Condeelis, J.; Pollard, J.W. Macrophages promote collagen fibrillogenesis around terminal end buds of the developing mammary gland. Dev. Dyn. 2006, 235, 3222–3229. [Google Scholar]
- Etich, J.; Bergmeier, V.; Frie, C.; Kreft, S.; Bengestrate, L.; Eming, S.; Mauch, C.; Eckes, B.; Ulus, H.; Lund, F.E.; et al. PECAM1+/Sca1+/CD38+ Vascular Cells Transform into Myofibroblast-Like Cells in Skin Wound Repair. PLoS ONE 2013, 8, e53262. [Google Scholar]
- Pöschl, E.; Schlötzer-Schrehardt, U.; Brachvogel, B.; Saito, K.; Ninomiya, Y.; Mayer, U. Collagen IV is essential for basement membrane stability but dispensable for initiation of its assembly during early development. Development 2004, 131, 1619–1628. [Google Scholar] [Green Version]
- Baer, C.; Squadrito, M.L.; Iruela-Arispe, M.L.; De Palma, M. Reciprocal interactions between endothelial cells and macrophages in angiogenic vascular niches. Exp. Cell Res. 2013, 319, 1626–1634. [Google Scholar]
- Bahr, J.C.; Weiss, S.J. Macrophage-Dependent Trafficking and Remodeling of the Basement Membrane-Interstitial Matrix Interface. BioRxiv 2018. [Google Scholar] [CrossRef]
- Iorio, V.; Troughton, L.D.; Hamill, K.J. Laminins: Roles and Utility in Wound Repair. Adv. Wound Care 2015, 4, 250. [Google Scholar]
- Pouliot, N.; Saunders, N.A.; Kaur, P. Laminin 10/11: An alternative adhesive ligand for epidermal keratinocytes with a functional role in promoting proliferation and migration. Exp. Dermatol. 2002, 11, 387–397. [Google Scholar]
- Aumailley, M.; Smyth, N. The role of laminins in basement membrane function. J. Anat. 1998, 193, 1–21. [Google Scholar]
- Ramos-Lewis, W.; LaFever, K.S.; Page-McCaw, A. A scar-like lesion is apparent in basement membrane after wound repair in vivo. Matrix Biol. 2018, 74, 101–120. [Google Scholar]
- Koivisto, L.; Heino, J.; Häkkinen, L.; Larjava, H. Integrins in Wound Healing. Adv. Wound Care 2014, 3, 762–783. [Google Scholar] [Green Version]
- Schiavinato, A.; Keene, D.R.; Wohl, A.P.; Corallo, D.; Colombatti, A.; Wagener, R.; Paulsson, M.; Bonaldo, P.; Sengle, G. Targeting of EMILIN-1 and EMILIN-2 to Fibrillin Microfibrils Facilitates their Incorporation into the Extracellular Matrix. J. Investig. Dermatol. 2016, 136, 1150–1160. [Google Scholar] [Green Version]
- Hayes, A.J.; Shu, C.C.; Lord, M.S.; Little, C.B.; Whitelock, J.M.; Melrose, J. Pericellular colocalisation and interactive properties of type VI collagen and perlecan in the intervertebral disc. Eur. Cell. Mater. 2016, 32, 40–57. [Google Scholar] [Green Version]
- Hayes, A.J.; Lord, M.S.; Smith, S.M.; Smith, M.M.; Whitelock, J.M.; Weiss, A.S.; Melrose, J. Colocalization in vivo and association in vitro of perlecan and elastin. Histochem. Cell Biol. 2011, 136, 437–454. [Google Scholar]
- Villar, M.J.; Hassell, J.R.; Brandan, E. Interaction of skeletal muscle cells with collagen type IV is mediated by perlecan associated with the cell surface. J. Cell. Biochem. 1999, 75, 665–674. [Google Scholar]
- Smith, S.M.; Melrose, J. Type XI collagen–perlecan–HS interactions stabilise the pericellular matrix of annulus fibrosus cells and chondrocytes providing matrix stabilisation and homeostasis. J. Mol. Histol. 2019, 50, 285–294. [Google Scholar]
- Ishihara, J.; Ishihara, A.; Fukunaga, K.; Sasaki, K.; White, M.J.V.; Briquez, P.S.; Hubbell, J.A. Laminin heparin-binding peptides bind to several growth factors and enhance diabetic wound healing. Nat. Commun. 2018, 9, 2163. [Google Scholar]
- Barrientos, S.; Stojadinovic, O.; Golinko, M.S.; Brem, H.; Tomic-Canic, M. PERSPECTIVE ARTICLE: Growth factors and cytokines in wound healing. Wound Repair Regen. 2008, 16, 585–601. [Google Scholar]
- Serafini, T.; Kennedy, T.E.; Gaiko, M.J.; Mirzayan, C.; Jessell, T.M.; Tessier-Lavigne, M. The netrins define a family of axon outgrowth-promoting proteins homologous to C. elegans UNC-6. Cell 1994, 78, 409–424. [Google Scholar]
- Mehlen, P.; Delloye-Bourgeois, C.; Chédotal, A. Novel roles for Slits and netrins: Axon guidance cues as anticancer targets? Nat. Rev. Cancer 2011, 11, 188–197. [Google Scholar]
- Grandin, M.; Meier, M.; Delcros, J.G.; Nikodemus, D.; Reuten, R.; Patel, T.R.; Goldschneider, D.; Orriss, G.; Krahn, N.; Boussouar, A.; et al. Structural Decoding of the Netrin-1/UNC5 Interaction and its Therapeutical Implications in Cancers. Cancer Cell 2016, 29, 173–185. [Google Scholar] [Green Version]
- Zhang, Y.; Chen, P.; Di, G.; Qi, X.; Zhou, Q.; Gao, H. Netrin-1 promotes diabetic corneal wound healing through molecular mechanisms mediated via the adenosine 2B receptor. Sci. Rep. 2018, 8, 5994. [Google Scholar]
- Colombelli, C.; Palmisano, M.; Eshed-Eisenbach, Y.; Zambroni, D.; Pavoni, E.; Ferri, C.; Saccucci, S.; Nicole, S.; Soininen, R.; McKee, K.K.; et al. Perlecan is recruited by dystroglycan to nodes of Ranvier and binds the clustering molecule gliomedin. J. Cell Biol. 2015, 208, 313–329. [Google Scholar] [Green Version]
- Noonan, D.M.; Fulle, A.; Valente, P.; Cai, S.; Horigan, E.; Sasaki, M.; Yamada, Y.; Hassell, J.R. The complete sequence of perlecan, a basement membrane heparan sulfate proteoglycan, reveals extensive similarity with laminin A chain, low density lipoprotein-receptor, and the neural cell adhesion molecule. J. Biol. Chem. 1991, 266, 22939–22947. [Google Scholar]
- Hayes, A.; Sugahara, K.; Farrugia, B.; Whitelock, J.M.; Caterson, B.; Melrose, J. Biodiversity of CS–proteoglycan sulphation motifs: Chemical messenger recognition modules with roles in information transfer, control of cellular behaviour and tissue morphogenesis. Biochem. J. 2018, 475, 587–620. [Google Scholar]
- Smith, M.M.; Melrose, J. Proteoglycans in Normal and Healing Skin. Adv. Wound Care 2015, 4, 152–173. [Google Scholar] [Green Version]
- Melrose, J. Glycosaminoglycans in Wound Healing. Bone Tissue Regen. Insights 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Miao, L.; Wang, Y.; Xia, H.; Yao, C.; Cai, H.; Song, Y. SPOCK1 is a novel transforming growth factor-β target gene that regulates lung cancer cell epithelial-mesenchymal transition. Biochem. Biophys. Res. Commun. 2013, 440, 792–797. [Google Scholar]
- Haensel, D.; Dai, X. Epithelial-to-mesenchymal transition in cutaneous wound healing: Where we are and where we are heading. Dev. Dyn. 2018, 247, 473–480. [Google Scholar]
- Hancock, G.E.; Kaplan, G.; Cohn, Z.A. Keratinocyte growth regulation by the products of immune cells. J. Exp. Med. 1988, 168, 1395–1402. [Google Scholar]
- Berger, E.A.; McClellan, S.A.; Barrett, R.P.; Hazlett, L.D. Testican-1 promotes resistance against Pseudomonas aeruginosa-induced keratitis through regulation of MMP-2 expression and activation. Investig. Ophthalmol. Vis. Sci. 2011, 52, 5339–5346. [Google Scholar]
- Yanagisawa, H.; Davis, E.C.; Starcher, B.C.; Ouchi, T.; Yanagisawa, M.; Richardson, J.A.; Olson, E.N. Fibulin-5 is an elastin-binding protein essential for elastic fibre development in vivo. Nature 2002, 415, 168–171. [Google Scholar]
- Nakamura, T.; Lozano, P.R.; Ikeda, Y.; Iwanaga, Y.; Hinek, A.; Minamisawa, S.; Cheng, C.-F.; Kobuke, K.; Dalton, N.; Takada, Y.; et al. Fibulin-5/DANCE is essential for elastogenesis in vivo. Nature 2002, 415, 171–175. [Google Scholar]
- Nakamura, T.; Ruiz-Lozano, P.; Lindner, V.; Yabe, D.; Taniwaki, M.; Furukawa, Y.; Kobuke, K.; Tashiro, K.; Lu, Z.; Andon, N.L.; et al. DANCE, a novel secreted RGD protein expressed in developing, atherosclerotic, and balloon-injured arteries. J. Biol. Chem. 1999, 274, 22476–22483. [Google Scholar]
- Albig, A.R.; Schiemann, W.P. Fibulin-5 Antagonizes Vascular Endothelial Growth Factor (VEGF) Signaling and Angiogenic Sprouting by Endothelial Cells. DNA Cell Biol. 2004, 23, 367–379. [Google Scholar]
- McCarty, S.M.; Percival, S.L. Proteases and Delayed Wound Healing. Adv. Wound Care 2013, 2, 438–447. [Google Scholar]
- Lim, N.H.; Kashiwagi, M.; Visse, R.; Jones, J.; Enghild, J.J.; Brew, K.; Nagase, H. Reactive-site mutants of N-TIMP-3 that selectively inhibit ADAMTS-4 and ADAMTS-5: Biological and structural implications. Biochem. J. 2010, 431, 113–122. [Google Scholar]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 786–801. [Google Scholar]
- Bekhouche, M.; Leduc, C.; Dupont, L.; Janssen, L.; Delolme, F.; Vadon-Le Goff, S.; Smargiasso, N.; Baiwir, D.; Mazzucchelli, G.; Zanella-Cleon, I.; et al. Determination of the substrate repertoire of ADAMTS2, 3, and 14 significantly broadens their functions and identifies extracellular matrix organization and TGF-β signaling as primary targets. FASEB J. 2016, 30, 1741–1756. [Google Scholar]
- Ricard-Blum, S.; Salza, R. Matricryptins and matrikines: Biologically active fragments of the extracellular matrix. Exp. Dermatol. 2014, 23, 457–463. [Google Scholar]
- Tran, K.T.; Lamb, P.; Deng, J.-S. Matrikines and matricryptins: Implications for cutaneous cancers and skin repair. J. Dermatol. Sci. 2005, 40, 11–20. [Google Scholar]
- Heljasvaara, R.; Nyberg, P.; Luostarinen, J.; Parikka, M.; Heikkilä, P.; Rehn, M.; Sorsa, T.; Salo, T.; Pihlajaniemi, T. Generation of biologically active endostatin fragments from human collagen XVIII by distinct matrix metalloproteases. Exp. Cell Res. 2005, 307, 292–304. [Google Scholar]
- Felbor, U. Secreted cathepsin L generates endostatin from collagen XVIII. EMBO J. 2000, 19, 1187–1194. [Google Scholar]
- Wen, W.; Moses, M.A.; Wiederschain, D.; Arbiser, J.L.; Folkman, J. The generation of endostatin is mediated by elastase. Cancer Res. 1999, 59, 6052–6056. [Google Scholar]
- Park, J.; Scherer, P.E. Adipocyte-derived endotrophin promotes malignant tumor progression. J. Clin. Investig. 2012, 122, 4243–4256. [Google Scholar] [Green Version]
- Bu, D.; Crewe, C.; Kusminski, C.M.; Gordillo, R.; Ghaben, A.L.; Kim, M.; Park, J.; Deng, H.; Xiong, W.; Liu, X.-Z.; et al. Human endotrophin as a driver of malignant tumor growth. JCI Insight 2019, 4. [Google Scholar] [CrossRef]
- Heumüller, S.E.; Talantikite, M.; Napoli, M.; Armengaud, J.; Mörgelin, M.; Hartmann, U.; Sengle, G.; Paulsson, M.; Moali, C.; Wagener, R. C-terminal proteolysis of the collagen VI α3 chain by BMP-1 and proprotein convertase(s) releases endotrophin in fragments of different sizes. J. Biol. Chem. 2019, 294, 13769–13780. [Google Scholar]
- Mundel, T.M.; Yliniemi, A.M.; Maeshima, Y.; Sugimoto, H.; Kieran, M.; Kalluri, R. Type IV collagen α6 chain-derived noncollagenous domain 1 (α6(IV)NC1) inhibits angiogenesis and tumor growth. Int. J. Cancer 2008, 122, 1738–1744. [Google Scholar]
- Maeshima, Y.; Colorado, P.C.; Torre, A.; Holthaus, K.A.; Grunkemeyer, J.A.; Ericksen, M.B.; Hopfer, H.; Xiao, Y.; Stillman, I.E.; Kalluri, R. Distinct antitumor properties’ of a type IV collagen domain derived from basement membrane. J. Biol. Chem. 2000, 275, 21340–21348. [Google Scholar]
- Pasco, S.; Ramont, L.; Venteo, L.; Pluot, M.; Maquart, F.X.; Monboisse, J.C. In vivo overexpression of tumstatin domains by tumor cells inhibits their invasive properties in a mouse melanoma model. Exp. Cell Res. 2004, 301, 251–265. [Google Scholar]
- Kamphaus, G.D.; Colorado, P.C.; Panka, D.J.; Hopfer, H.; Ramchandran, R.; Torre, A.; Maeshima, Y.; Mier, J.W.; Sukhatme, V.P.; Kalluri, R. Canstatin, a novel matrix-derived inhibitor of angiogenesis and tumor growth. J. Biol. Chem. 2000, 275, 1209–1215. [Google Scholar]
- Karagiannis, E.D.; Popel, A.S. Identification of novel short peptides derived from the α4, α5, and α6 fibrils of type IV collagen with anti-angiogenic properties. Biochem. Biophys. Res. Commun. 2007, 354, 434–439. [Google Scholar]
- Colorado, P.C.; Torre, A.; Kamphaus, G.; Maeshima, Y.; Hopfer, H.; Takahashi, K.; Volk, R.; Zamborsky, E.D.; Herman, S.; Sarkar, P.K.; et al. Anti-angiogenic cues from vascular basement membrane collagen. Cancer Res. 2000, 60, 2520–2526. [Google Scholar]
- Sage, E.H.; Reed, M.; Funk, S.E.; Truong, T.; Steadele, M.; Puolakkainen, P.; Maurice, D.H.; Bassuk, J.A. Cleavage of the matricellular protein SPARC by matrix metalloproteinase 3 produces polypeptides that influence angiogenesis. J. Biol. Chem. 2003, 278, 37849–37857. [Google Scholar]
- Rousselle, P.; Carulli, S.; Chajra, H.; Dayan, G.; Pin, D.; Herbage, B. The syndecan binding sequence KKLRIKSKEK in laminin alpha3 LG4 domain promotes epidermal repair. Eur. J. Dermatol. 2013. Available online: https://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/pubmed/23567164/ (accessed on 23 June 2019).
- Pathan, M.; Keerthikumar, S.; Ang, C.S.; Gangoda, L.; Quek, C.Y.J.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A.; et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar]
- Etich, J.; Bergmeier, V.; Pitzler, L.; Brachvogel, B. Identification of a reference gene for the quantification of mRNA and miRNA expression during skin wound healing. Connect. Tissue Res. 2017, 58, 196–207. [Google Scholar]
- Maertens, B.; Hopkins, D.; Franzke, C.W.; Keene, D.R.; Bruckner-Tuderman, L.; Greenspan, D.S.; Koch, M. Cleavage and oligomerization of gliomedin, a transmembrane collagen required for node of Ranvier formation. J. Biol. Chem. 2007, 282, 10647–10659. [Google Scholar]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Etich, J.; Koch, M.; Wagener, R.; Zaucke, F.; Fabri, M.; Brachvogel, B. Gene Expression Profiling of the Extracellular Matrix Signature in Macrophages of Different Activation Status: Relevance for Skin Wound Healing. Int. J. Mol. Sci. 2019, 20, 5086. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205086
Etich J, Koch M, Wagener R, Zaucke F, Fabri M, Brachvogel B. Gene Expression Profiling of the Extracellular Matrix Signature in Macrophages of Different Activation Status: Relevance for Skin Wound Healing. International Journal of Molecular Sciences. 2019; 20(20):5086. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205086
Chicago/Turabian StyleEtich, Julia, Manuel Koch, Raimund Wagener, Frank Zaucke, Mario Fabri, and Bent Brachvogel. 2019. "Gene Expression Profiling of the Extracellular Matrix Signature in Macrophages of Different Activation Status: Relevance for Skin Wound Healing" International Journal of Molecular Sciences 20, no. 20: 5086. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205086