Mutation of ONAC096 Enhances Grain Yield by Increasing Panicle Number and Delaying Leaf Senescence during Grain Filling in Rice
Abstract
:1. Introduction
2. Results
2.1. Characterization of ONAC096
2.2. ONAC096 is Upregulated During Leaf Senescence
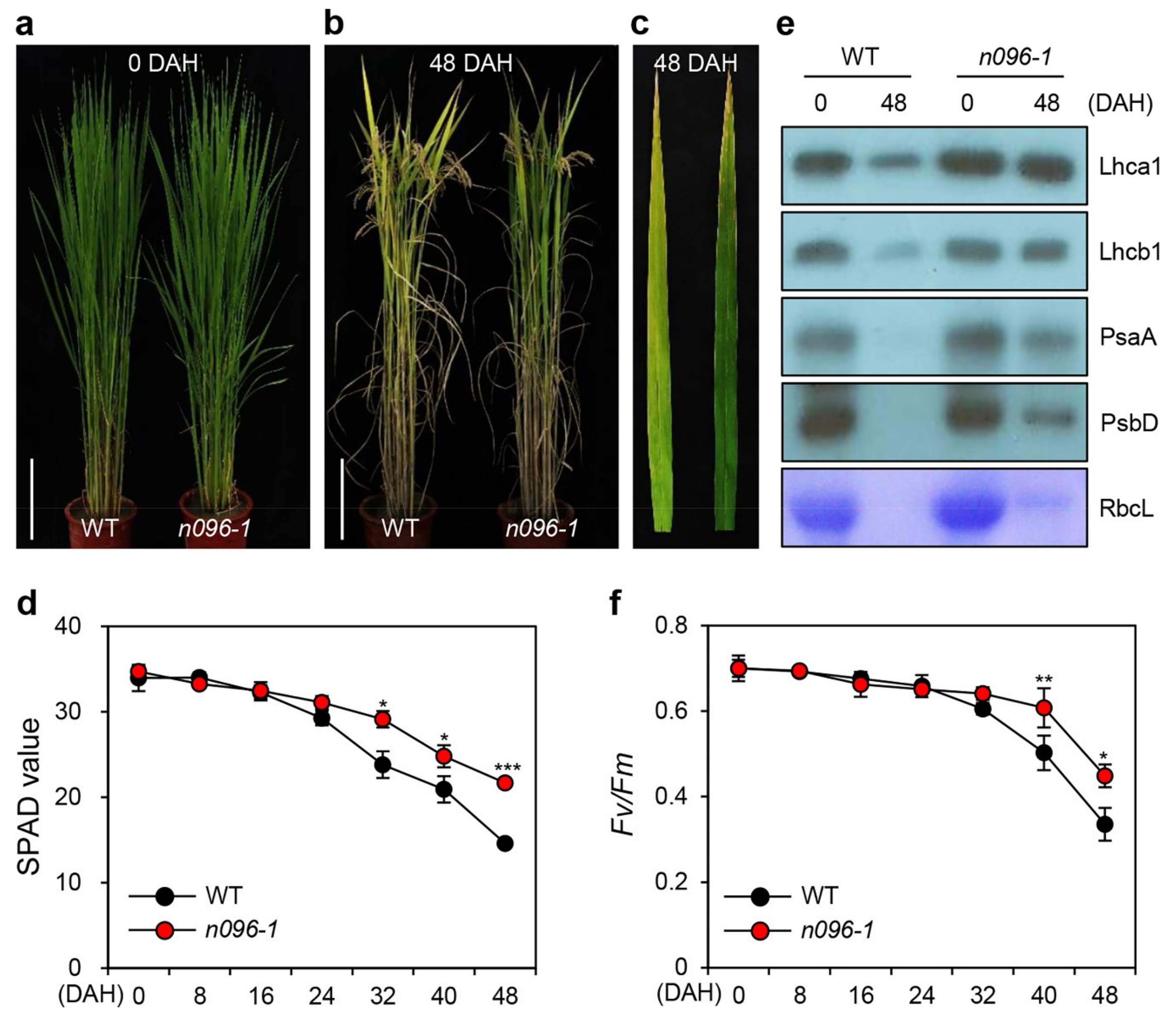
2.3. The onac096 Mutation Delays Leaf Yellowing during Natural and Dark-Induced Senescence
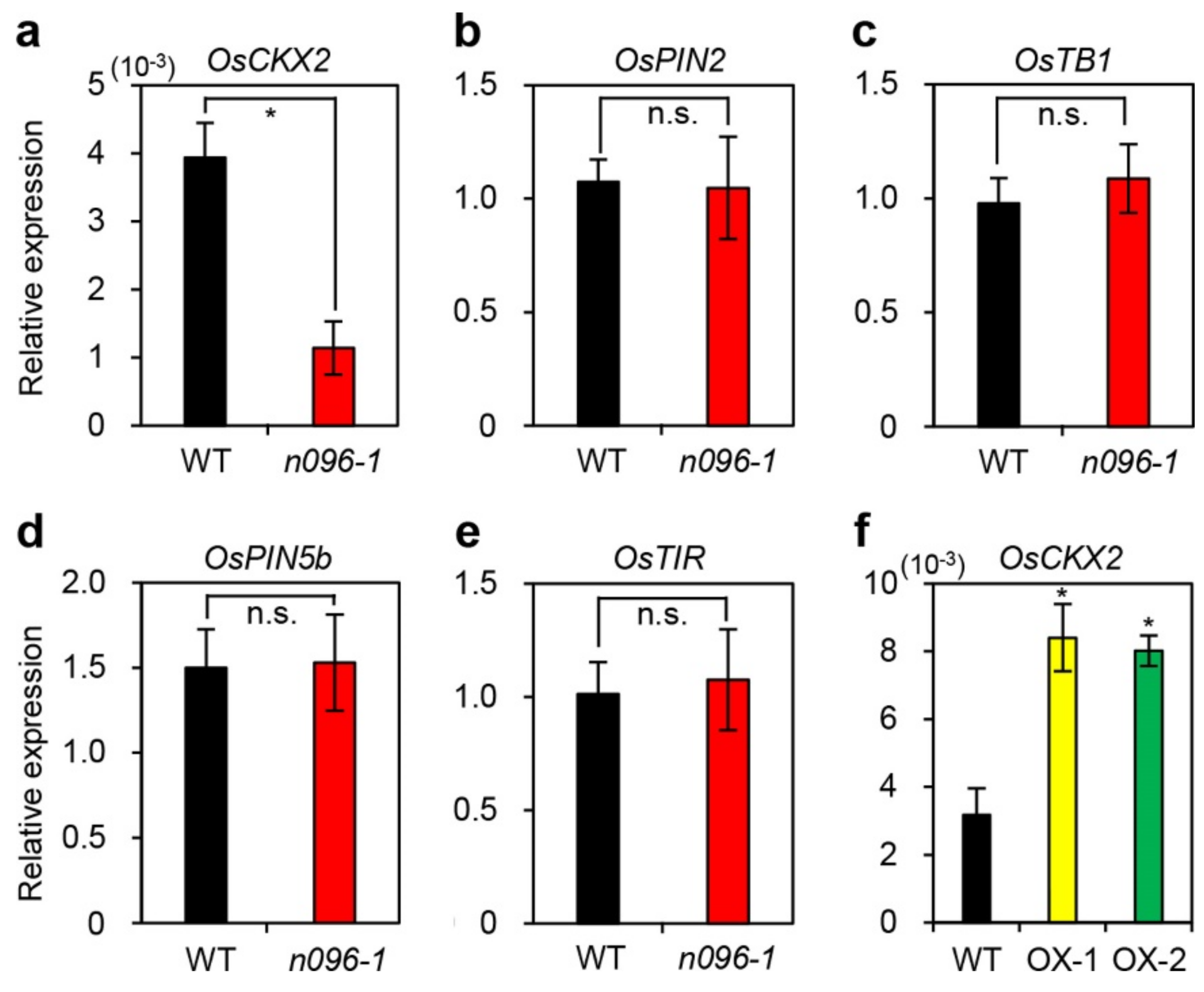
2.4. Mutation of ONAC096 Improves Grain Yield by Increasing the Number of Panicles Per Plant
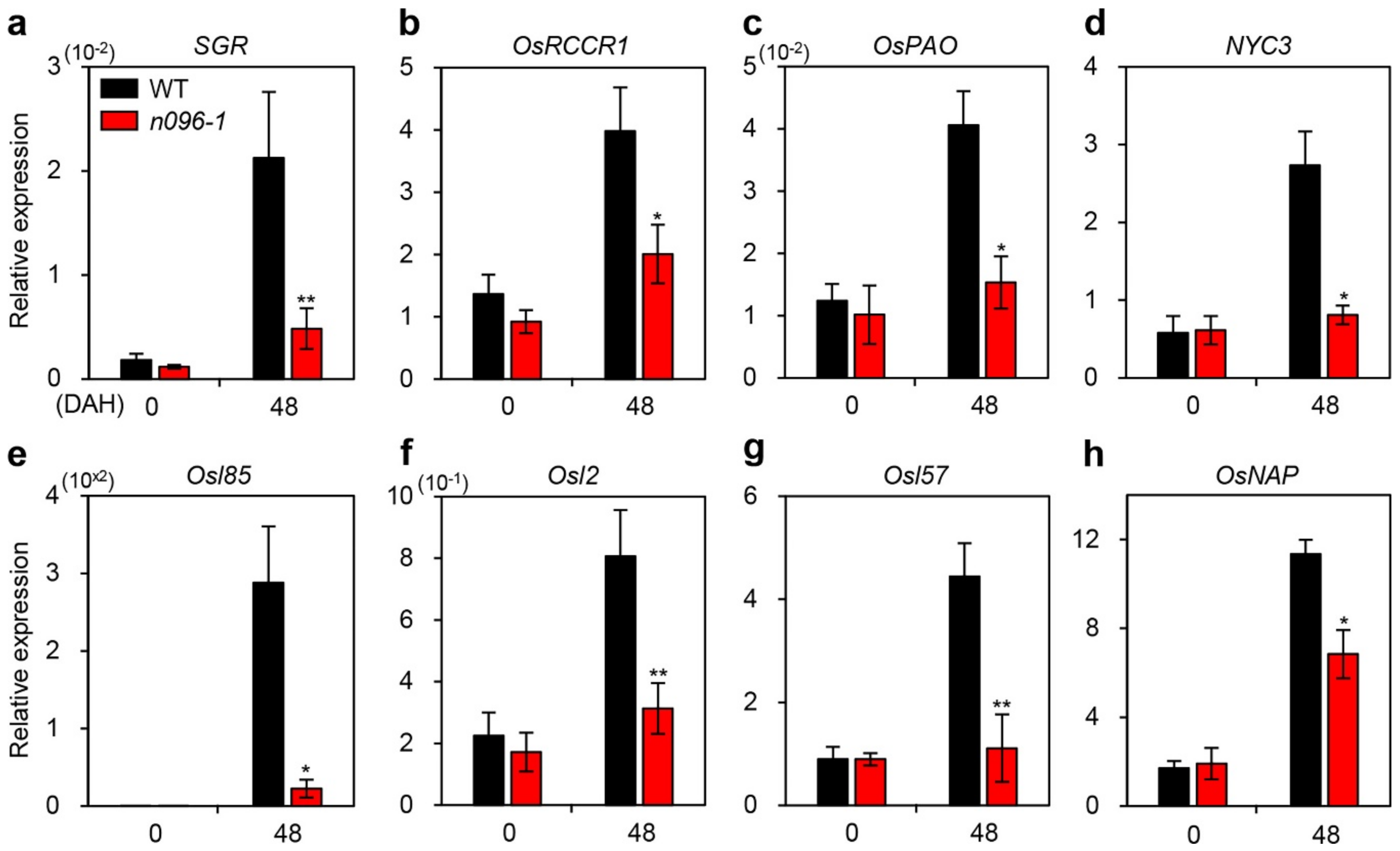
2.5. ONAC096 Upregulates CDG and SAG Expression During Leaf Senescence
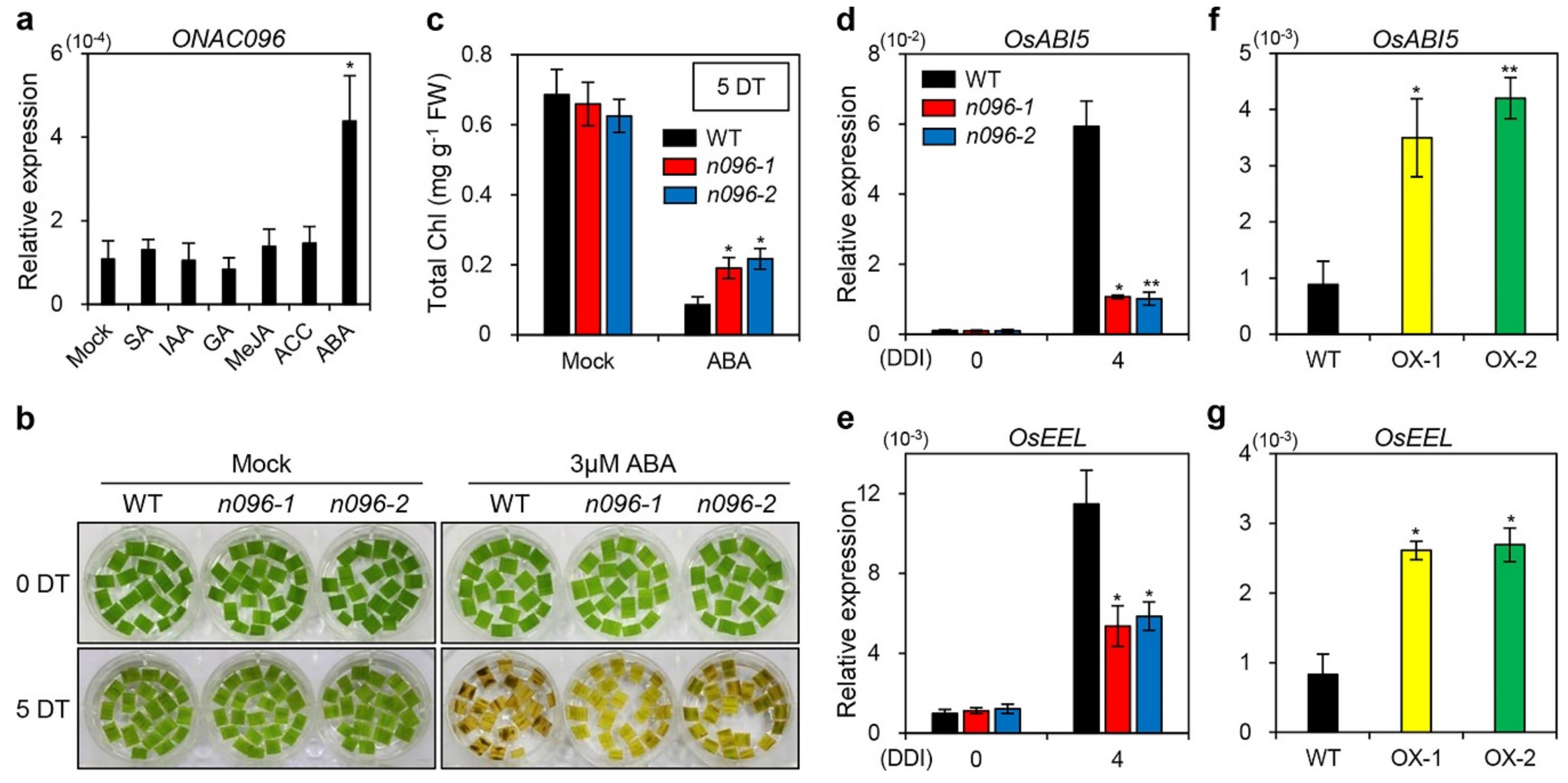
2.6. ONAC096 Mediates ABA-Induced Leaf Senescence
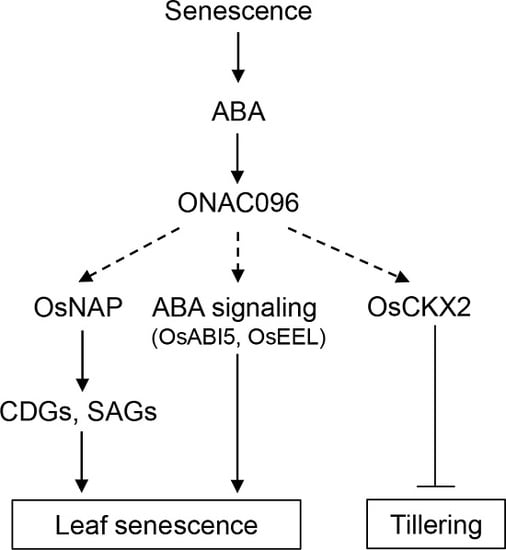
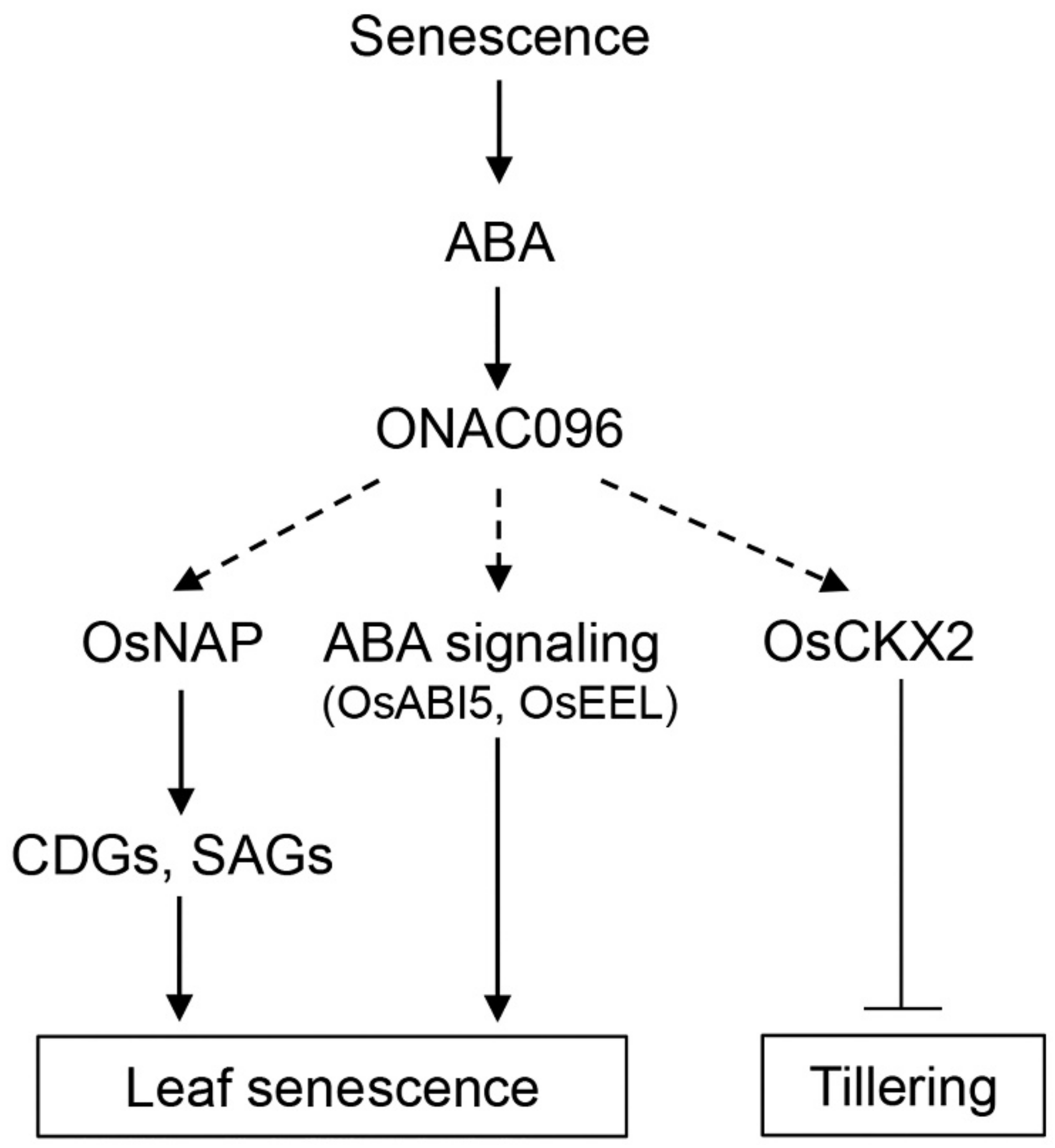
3. Discussion
3.1. Effects of ONAC096 on Leaf Senescence and Tillering
3.2. Involvement of ONAC096 in ABA-Induced Leaf Senescence
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Experimental Treatments
4.2. Determination of Photosynthetic Activity, Total Chlorophyll Content, and SPAD Value
4.3. RT-qPCR Analysis
4.4. Plasmid Construction and Rice Transformation
4.5. SDS-PAGE and Immunoblot Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| WT | wild type |
| NLD | natural long day |
| LD | long day |
| CDGs | chlorophyll degradation genes |
| SAGs | senescence-associated genes |
| DIS | dark-induced senescence |
| DDI | day(s) of dark incubation |
| DAS | day(s) after seeding |
| DAH | day(s) after heading |
| DT | day(s) after treatment |
References
- Khush, G.S. Origin, dispersal, cultivation and variation of rice. Plant Mol. Biol. 1997, 35, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Numaguchi, K.; Miura, K.; Yoshida, K.; Thanh, P.T.; Htun, T.M.; Yamasaki, M.; Komeda, N.; Matsumoto, T.; Terauchi, R.; et al. OsLG1 regulates a closed panicle trait in domesticated rice. Nat. Genet. 2013, 45, 462–465. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Liu, H.; Zhou, T.; Gu, B.; Huang, X.; Shangguan, Y.; Zhu, J.; Li, Y.; Zhao, Y.; Wang, Y.; et al. An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell 2013, 25, 3360–3376. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Tan, L.; Fu, Y.; Liu, F.; Cai, H.; Xie, D.; Wu, F.; Wu, J.; Matsumoto, T.; Sun, C. Genetic control of inflorescence architecture during rice domestication. Nat. Commun. 2013, 4, 2200. [Google Scholar] [CrossRef]
- Khush, G. Productivity improvements in rice. Nutr. Rev. 2003, 61, S114–S116. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.Q.; Zhu, J.; He, C.X.; Benmoussa, M.; Wu, P. Quantitative trait loci analysis for the developmental behavior of tiller number in rice (Oryza sativa L.). Theor. Appl. Genet. 1998, 97, 267–274. [Google Scholar] [CrossRef]
- Liang, W.; Shang, F.; Lin, Q.; Lou, C.; Zhang, J. Tillering and panicle branching genes in rice. Gene 2014, 537, 1–5. [Google Scholar] [CrossRef]
- Takeda, T.; Suwa, Y.; Suzuki, M.; Kitano, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Matsuoka, M.; Ueguchi, C. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 2003, 33, 513–520. [Google Scholar] [CrossRef]
- Lim, P.O.; Kim, H.J.; Nam, H.G. Leaf senescence. Annu. Rev. Plant Biol. 2007, 58, 115–136. [Google Scholar] [CrossRef]
- Morita, R.; Sato, Y.; Masuda, Y.; Nishimura, M.; Kusaba, M. Defect in non-yellow coloring 3, an α/β hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J. 2009, 59, 940–952. [Google Scholar] [CrossRef]
- Tang, Y.; Li, M.; Chen, Y.; Wu, P.; Wu, G.; Jiang, H. Knockdown of OsPAO and OsRCCR1 cause different plant death phenotypes in rice. J. Plant Physiol. 2011, 168, 1952–1959. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Yu, J.-W.; Park, J.-S.; Li, J.; Yoo, S.-C.; Lee, N.-Y.; Lee, S.-K.; Jeong, S.-W.; Seo, H.S.; Koh, H.-J.; et al. The senescence-induced staygreen protein regulates chlorophyll degradation. Plant Cell 2007, 19, 1649–1664. [Google Scholar] [CrossRef] [PubMed]
- Shimoda, Y.; Ito, H.; Tanaka, A. Arabidopsis STAY-GREEN, mendel’s green cotyledon gene, encodes magnesium-dechelatase. Plant Cell 2016, 28, 2147–2160. [Google Scholar] [CrossRef] [PubMed]
- Mao, C.; Lu, S.; Lv, B.; Zhang, B.; Shen, J.; He, J.; Luo, L.; Xi, D.; Chen, X.; Ming, F. A Rice NAC transcription factor promotes leaf senescence via ABA biosynthesis. Plant Physiol. 2017, 174, 1747–1763. [Google Scholar] [CrossRef]
- Liang, C.; Wang, Y.; Zhu, Y.; Tang, J.; Hu, B.; Liu, L.; Ou, S.; Wu, H.; Sun, X.; Chu, J.; et al. OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 10013–10018. [Google Scholar] [CrossRef] [Green Version]
- Sperotto, R.A.; Ricachenevsky, F.K.; Duarte, G.L.; Boff, T.; Lopes, K.L.; Sperb, E.R.; Grusak, M.A.; Fett, J.P. Identification of up-regulated genes in flag leaves during rice grain filling and characterization of OsNAC5, a new ABA-dependent transcription factor. Planta 2009, 230, 985–1002. [Google Scholar] [CrossRef]
- Nakashima, K.; Tran, L.-S.P.; Van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef]
- Woodward, A.W.; Bartel, B. Auxin: Regulation, action, and interaction. Ann. Bot. 2005, 95, 707–735. [Google Scholar] [CrossRef]
- Chen, Y.; Fan, X.; Song, W.; Zhang, Y.; Xu, G. Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol. J. 2012, 10, 139–149. [Google Scholar] [CrossRef]
- Lu, G.; Coneva, V.; Casaretto, J.A.; Ying, S.; Mahmood, K.; Liu, F.; Nambara, E.; Bi, Y.-M.; Rothstein, S.J. OsPIN5b modulates rice (Oryza sativa) plant architecture and yield by changing auxin homeostasis, transport and distribution. Plant J. 2015, 83, 913–925. [Google Scholar] [CrossRef]
- Xia, K.; Wang, R.; Ou, X.; Fang, Z.; Tian, C.; Duan, J.; Wang, Y.; Zhang, M. OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS ONE 2012, 7, e30039. [Google Scholar] [CrossRef] [PubMed]
- Yeh, S.-Y.; Chen, H.-W.; Ng, C.-Y.; Lin, C.-Y.; Tseng, T.-H.; Li, W.-H.; Ku, M.S.B. Down-regulation of cytokinin oxidase 2 expression Increases tiller number and improves rice yield. Rice 2015, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Chandler, P.M.; Robertson, M. Gene expression regulated by abscisic acid and its relation to stress tolerance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1994, 45, 113–141. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Jibran, R.; Hunter, D.A.; P. Dijkwel, P. Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Mol. Biol. 2013, 82, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Even-Chen, Z.; Itai, C. The role of abscisic acid in senescence of detached tobacco leaves. Physiol. Plant 1975, 34, 97–100. [Google Scholar] [CrossRef]
- Gepstein, S.; Thimann, K.V. Changes in the abscisic acid content of oat leaves during senescence. Proc. Natl. Acad. Sci. USA 1980, 77, 2050–2053. [Google Scholar] [CrossRef] [Green Version]
- Philosoph-Hadas, S.; Hadas, E.; Aharoni, N. Characterization and use in ELISA of a new monoclonal antibody for quantitation of abscisic acid in senescing rice leaves. Plant Growth Regul. 1993, 12, 71–78. [Google Scholar] [CrossRef]
- He, P.; Osaki, M.; Takebe, M.; Shinano, T.; Wasaki, J. Endogenous hormones and expression of senescence-related genes in different senescent types of maize. J. Exp. Bot. 2005, 56, 1117–1128. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.-F.; Qiu, K.; Ren, G.-D.; Zhu, Y.; Kuai, B.-K. A pleiotropic phenotype is associated with altered endogenous hormone balance in the developmentally stunted mutant (dsm1). J. Plant Biol. 2010, 53, 79–87. [Google Scholar] [CrossRef]
- Tan, B.-C.; Joseph, L.M.; Deng, W.-T.; Liu, L.; Li, Q.-B.; Cline, K.; McCarty, D.R. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003, 35, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Guo, Y.; Liu, Y.; Zhang, F.; Wang, Z.; Wang, H.; Wang, F.; Li, D.; Mao, D.; Luan, S.; et al. 9-cis-epoxycarotenoid dioxygenase 3 regulates plant growth and enhances multi-abiotic Stress tolerance in rice. Front. Plant Sci. 2018, 9, 162. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.-D.; Seo, P.J.; Yoon, H.-K.; Park, C.-M. The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 2011, 23, 2155–2168. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Chung, K.M.; Woo, H.R. Three positive regulators of leaf senescence in Arabidopsis, ORE1, ORE3 and ORE9, play roles in crosstalk among multiple hormone-mediated senescence pathways. Genes Genom. 2011, 33, 373–381. [Google Scholar] [CrossRef]
- Sun, L.; Huang, L.; Hong, Y.; Zhang, H.; Song, F.; Li, D. Comprehensive analysis suggests overlapping expression of rice ONAC transcription factors in abiotic and biotic stress responses. Int. J. Mol. Sci. 2015, 16, 4306–4326. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; You, J.; Xie, K.; Xie, W.; Xiong, L. Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genom. 2008, 280, 547–563. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, Y.; Piao, W.; Lim, J.-H.; Han, S.-H.; Kim, Y.-S.; An, G.; Paek, N.-C. Rice ONAC106 inhibits leaf senescence and increases salt tolerance and tiller angle. Plant Cell Physiol. 2015, 56, 2325–2339. [Google Scholar] [CrossRef]
- El Mannai, Y.; Akabane, K.; Hiratsu, K.; Satoh-Nagasawa, N.; Wabiko, H. The NAC transcription factor gene OsY37 (ONAC011) promotes leaf senescence and accelerates heading time in rice. Int. J. Mol. Sci. 2017, 18, 2165. [Google Scholar] [CrossRef]
- Thomas, H.; Smart, C.M. Crops that stay green. Ann. Appl. Biol. 1993, 123, 193–219. [Google Scholar] [CrossRef]
- Moldenhauer, K.A.K.; Gibbons, J.H. Rice morphology and development. In Rice: Origin, History, Technology, and Production; Smith, C.W., Dilday, R.H., Eds.; Wiley: Hoboken, NJ, USA, 2003; pp. 103–128. ISBN 978–0-471-34516-9. [Google Scholar]
- Badshah, M.A.; Naimei, T.; Zou, Y.; Ibrahim, M.; Wang, K. Yield and tillering response of super hybrid rice Liangyoupeijiu to tillage and establishment methods. Crop J. 2014, 2, 79–86. [Google Scholar] [CrossRef]
- Lee, R.H.; Wang, C.H.; Huang, L.T.; Chen, S.C. Leaf senescence in rice plants: Cloning and characterization of senescence up-regulated genes. J. Exp. Bot. 2001, 52, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhou, C. Signal transduction in leaf senescence. Plant Mol. Biol. 2013, 82, 539–545. [Google Scholar] [CrossRef] [PubMed]
- Zou, M.; Guan, Y.; Ren, H.; Zhang, F.; Chen, F. A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol. Biol. 2008, 66, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.R.; Shin, M.; Shen, J.Q. The wheat PKABA1-interacting factor TaABF1 mediates both abscisic acid-suppressed and abscisic acid-induced gene expression in bombarded aleurone cells. Plant Mol. Biol. 2008, 68, 93–103. [Google Scholar] [CrossRef]
- Lee, S.-H.; Sakuraba, Y.; Lee, T.; Kim, K.-W.; An, G.; Lee, H.Y.; Paek, N.-C. Mutation of Oryza sativa CORONATINE INSENSITIVE 1b (OsCOI1b) delays leaf senescence. J. Integr. Plant Biol. 2015, 57, 562–576. [Google Scholar] [CrossRef]
- Walulu, R.S.; Rosenow, D.T.; Wester, D.B.; Nguyen, H.T. Inheritance of the stay green trait in sorghum. Crop Sci. 1994, 34, 970. [Google Scholar] [CrossRef]
- Pierce, R.O.; Knowles, P.F.; Phillips, D. Inheritance of delayed leaf senescence in soybean. Crop Sci. 1984, 24, 515. [Google Scholar] [CrossRef]
- Gentinetta, E.; Ceppl, D.; Lepori, C.; Perico, G.; Motto, M.; Salamini, F. A major gene for delayed senescence in maize. Pattern of photosynthates accumulation and inheritance. Plant Breed. 1986, 97, 193–203. [Google Scholar] [CrossRef]
- Fang, Z.; Bouwkamp, J.C.; Solomos, T. Chlorophyllase activities and chlorophyll degradation during leaf senescence in non-yellowing mutant and wild type of Phaseolus vulgaris L. J. Exp. Bot. 1998, 49, 503–510. [Google Scholar]
- Spano, G.; Di Fonzo, N.; Perrotta, C.; Ronga, G.; Lawlor, D.W.; Napier, J.A.; Shewry, P.R. Physiological characterization of ‘stay green’ mutants in durum wheat. J. Exp. Bot. 2003, 54, 1415–1420. [Google Scholar] [CrossRef]
- Schittenhelm, S.; Menge-Hartmann, U.; Oldenburg, E. Photosynthesis, carbohydrate metabolism, and yield of phytochrome-B-overexpressing potatoes under different light regimes. Crop Sci. 2004, 44, 131. [Google Scholar] [CrossRef]
- Yoo, S.-C.; Cho, S.-H.; Zhang, H.; Paik, H.-C.; Lee, C.-H.; Li, J.; Yoo, J.-H.; Lee, B.-W.; Koh, H.-J.; Seo, H.S.; et al. Quantitative trait loci associated with functional stay-green SNU-SG1 in rice. Mol. Cells 2007, 24, 83–94. [Google Scholar] [PubMed]
- Thomas, H.; Howarth, C.J. Five ways to stay green. J. Exp. Bot. 2000, 51, 329–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Q.; Yuan, Z.; Chen, M.; Yin, C.; Luo, Z.; Zhao, X.; Liang, W.; Hu, J.; Zhang, D. Jasmonic acid regulates spikelet development in rice. Nat. Commun. 2014, 5, 3476. [Google Scholar] [CrossRef] [Green Version]
- Shim, Y.; Kang, K.; An, G.; Paek, N.-C. Rice DNA-binding one zinc finger 24 (OsDOF24) delays leaf senescence in a jasmonate-mediated pathway. Plant Cell Physiol. 2019. [Google Scholar] [CrossRef]
- Mae, T. Physiological nitrogen efficiency in rice: Nitrogen utilization, photosynthesis, and yield potential. Plant Soil 1997, 196, 201–210. [Google Scholar] [CrossRef]
- Miller, B.C.; Hill, J.E.; Roberts, S.R. Plant population effects on growth and yield in water-seeded rice. Agron. J. 1991, 83, 291. [Google Scholar] [CrossRef]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef]
- Bennett, M.J.; Marchant, A.; Green, H.G.; May, S.T.; Ward, S.P.; Millner, P.A.; Walker, A.R.; Schulz, B.; Feldmann, K.A. Arabidopsis AUX1 gene: A permease-like regulator of root gravitropism. Science 1996, 273, 948–950. [Google Scholar] [CrossRef]
- Friml, J.; Vieten, A.; Sauer, M.; Weijers, D.; Schwarz, H.; Hamann, T.; Offringa, R.; Jürgens, G. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature 2003, 426, 147–153. [Google Scholar] [CrossRef]
- Blilou, I.; Xu, J.; Wildwater, M.; Willemsen, V.; Paponov, I.; Friml, J.; Heidstra, R.; Aida, M.; Palme, K.; Scheres, B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 2005, 433, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Ashikari, M.; Sakakibara, H.; Lin, S.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.R.; Qian, Q.; Kitano, H.; Matsuoka, M. Cytokinin oxidase regulates rice grain production. Science 2005, 309, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Gan, S. Leaf senescence: Signals, execution, and regulation. Curr. Top. Dev. Biol. 2005, 71, 83–112. [Google Scholar] [PubMed]
- Noodén, L.D.; Singh, S.; Letham, D.S. Correlation of xylem sap cytokinin levels with monocarpic senescence in soybean. Plant Physiol. 1990, 93, 33–39. [Google Scholar] [CrossRef]
- Ambler, J.R.; Morgan, P.W.; Jordan, W.R. Amounts of zeatin and zeatin riboside in xylem sap of senscent and nonsenescent Sorghum. Crop Sci. 1992, 32, 411. [Google Scholar] [CrossRef]
- Buchanan-Wollaston, V.; Page, T.; Harrison, E.; Breeze, E.; Lim, P.O.; Nam, H.G.; Lin, J.-F.; Wu, S.-H.; Swidzinski, J.; Ishizaki, K.; et al. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 2005, 42, 567–585. [Google Scholar] [CrossRef]
- Breeze, E.; Harrison, E.; McHattie, S.; Hughes, L.; Hickman, R.; Hill, C.; Kiddle, S.; Kim, Y.-S.; Penfold, C.A.; Jenkins, D.; et al. High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 2011, 23, 873–894. [Google Scholar] [CrossRef]
- Tsai, Y.-C.; Weir, N.R.; Hill, K.; Zhang, W.; Kim, H.J.; Shiu, S.-H.; Schaller, G.E.; Kieber, J.J. Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol. 2012, 158, 1666–1684. [Google Scholar] [CrossRef]
- Joshi, R.; Sahoo, K.K.; Tripathi, A.K.; Kumar, R.; Gupta, B.K.; Pareek, A.; Singla-Pareek, S.L. Knockdown of an inflorescence meristem-specific cytokinin oxidase - OsCKX2 in rice reduces yield penalty under salinity stress condition. Plant Cell Environ. 2018, 41, 936–946. [Google Scholar] [CrossRef]
- Sakuraba, Y.; Jeong, J.; Kang, M.-Y.; Kim, J.; Paek, N.-C.; Choi, G. Phytochrome-interacting transcription factors PIF4 and PIF5 induce leaf senescence in Arabidopsis. Nat Commun. 2014, 5, 4636. [Google Scholar] [CrossRef] [Green Version]
- Jeong, D.-H.; An, S.; Kang, H.-G.; Moon, S.; Han, J.-J.; Park, S.; Lee, H.S.; An, K.; An, G. T-DNA insertional mutagenesis for activation tagging in rice. Plant Physiol. 2002, 130, 1636–1644. [Google Scholar] [CrossRef] [PubMed]
- Porra, R.J.; Thompson, W.A.; Kriedemann, P.E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys. Acta 1989, 975, 384–394. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.-S.; Lee, S.; Jung, K.-H.; Jun, S.-H.; Jeong, D.-H.; Lee, J.; Kim, C.; Jang, S.; Lee, S.; Yang, K.; et al. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 2000, 22, 561–570. [Google Scholar] [CrossRef] [PubMed]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, K.; Shim, Y.; Gi, E.; An, G.; Paek, N.-C. Mutation of ONAC096 Enhances Grain Yield by Increasing Panicle Number and Delaying Leaf Senescence during Grain Filling in Rice. Int. J. Mol. Sci. 2019, 20, 5241. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205241
Kang K, Shim Y, Gi E, An G, Paek N-C. Mutation of ONAC096 Enhances Grain Yield by Increasing Panicle Number and Delaying Leaf Senescence during Grain Filling in Rice. International Journal of Molecular Sciences. 2019; 20(20):5241. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205241
Chicago/Turabian StyleKang, Kiyoon, Yejin Shim, Eunji Gi, Gynheung An, and Nam-Chon Paek. 2019. "Mutation of ONAC096 Enhances Grain Yield by Increasing Panicle Number and Delaying Leaf Senescence during Grain Filling in Rice" International Journal of Molecular Sciences 20, no. 20: 5241. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20205241