Citrus Genetic Engineering for Disease Resistance: Past, Present and Future
Abstract
:1. Introduction
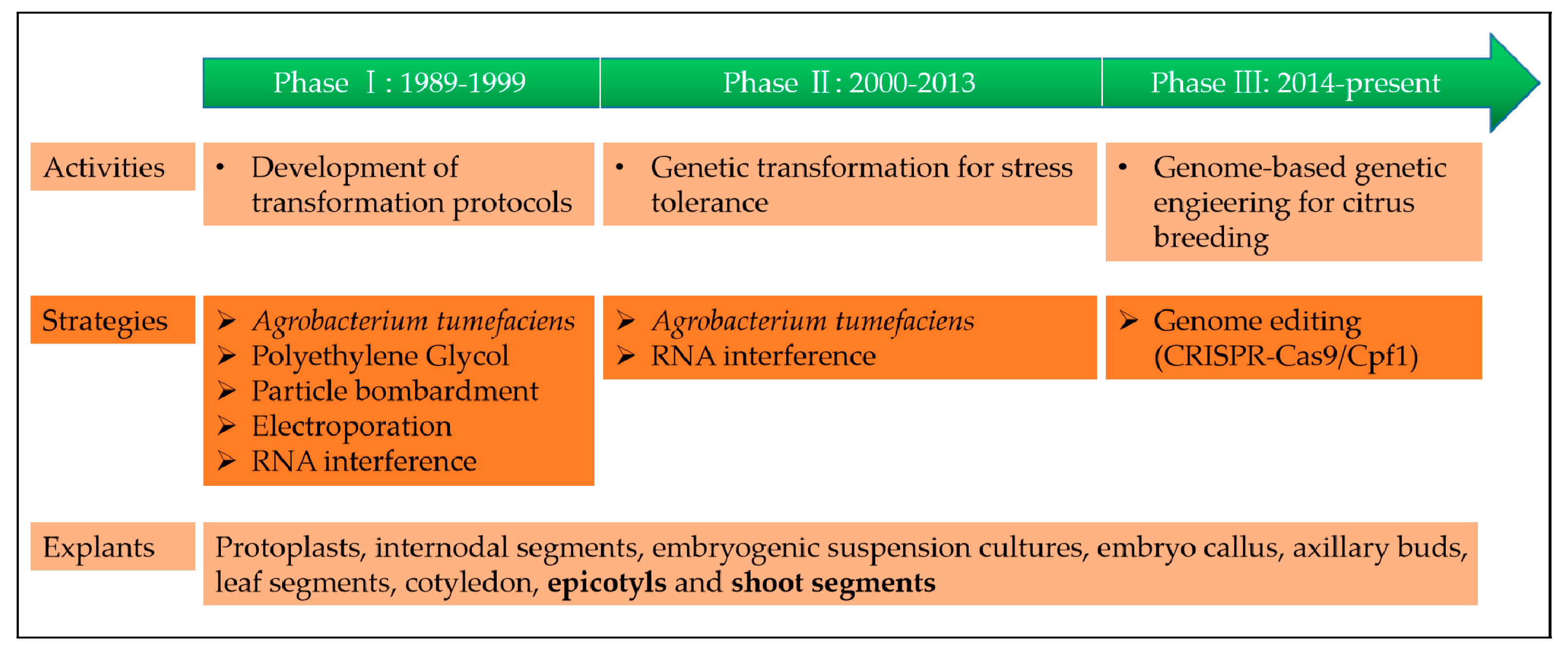
2. Genetic Engineering History of Citrus Crops
2.1. Phase I: Development of Transformation Protocols (1989–1999)
2.2. Phase II: Genetic Transformation for Stress Tolerance (2000–2013)
2.3. Phase III: Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CAS (CRISPR-Associated) Systems (2014–Present)
3. Advances in Transgenic Research for Bacterial Disease Resistance in Citrus
3.1. Transgenic Research Related to Canker Resistance
3.2. Transgenic Research for Huanglongbing Resistance
4. Advances in Transgenic Research for Fungal and Viral Disease Resistance in Citrus
5. Application of Genome-Editing Techniques in Disease-Resistance Breeding of Citrus
6. Genetic Approaches to Suppress Vector-Borne Bacterial/Viral Diseases in Citrus
7. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wu, G.A.; Terol, J.; Ibanez, V.; Lopez-Garcia, A.; Perez-Roman, E.; Borreda, C.; Domingo, C.; Tadeo, F.R.; Carbonell-Caballero, J.; Alonso, R.; et al. Genomics of the origin and evolution of Citrus. Nature 2018, 554, 311–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Febres, V.; Fisher, L.; Khalaf, A.; Moore, G.A. Citrus Transformation: Challenges and Prospects. In Genetic transformation; Alvarez, M., Ed.; IntechOpen: London, UK, 2011; pp. 101–122. [Google Scholar]
- Upadhyay, A. Transgenic Research in Fruit Crops. In Genetic Engineering of Horticultural Crops; Rout, G.R., Peter, K.V., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 63–87. [Google Scholar]
- Prabha, R.; Singh, D.P.; Verma, M.K. Microbial Interactions and Perspectives for Bioremediation of Pesticides in the Soils. In Plant-Microbe Interactions in Agro-Ecological Perspectives; Singh, D.P., Singh, H.B., Prabha, R., Eds.; Springer: Singapore, 2017; Volume 2, pp. 649–671. [Google Scholar]
- Gimenes-Fernandes, N.; Barbosa, J.C.; Ayres, A.; Massari, C. Plantas doentes não detectadas nas inspeções dificultam a erradicação do cancro cítrico. Summa Phytopathol. 2000, 26, 320–325. [Google Scholar] [CrossRef]
- Pan, X.L.; Dong, F.S.; Wu, X.H.; Xu, J.; Liu, X.G.; Zheng, Y.Q. Progress of the discovery, application, and control technologies of chemical pesticides in China. J. Integr. Agric. 2018, 18, 840–853. [Google Scholar] [CrossRef]
- Grosser, J.; Gmitter, F.; Tusa, N.; Chandler, J. Somatic hybrid plants from sexually incompatible woody species: Citrus reticulata and Citropsis gilletiana. Plant Cell Rep. 1990, 8, 656–659. [Google Scholar] [CrossRef] [PubMed]
- Vanloqueren, G.; Baret, P.V. How agricultural research systems shape a technological regime that develops genetic engineering but locks out agroecological innovations. Res. Policy 2009, 38, 971–983. [Google Scholar] [CrossRef]
- Gong, X.Q.; Liu, J.H. Genetic transformation and genes for resistance to abiotic and biotic stresses in Citrus and its related genera. Plant Cell Tiss. Org. 2012, 113, 137–147. [Google Scholar] [CrossRef]
- Orsenigo, L. The Emergence of Biotechnology: Institutions and Markets in Industrial Innovation; Pinter Publishers Ltd.: London, UK, 1989. [Google Scholar]
- Cohen, S.N.; Chang, A.C.; Boyer, H.W.; Helling, R.B. Construction of biologically functional bacterial plasmids in vitro. Proc. Natl. Acad. Sci. USA 1973, 70, 3240–3244. [Google Scholar] [CrossRef]
- Melchers, L.S.; Stuiver, M.H. Novel genes for disease-resistance breeding. Curr. Opin. Plant. Biol. 2000, 3, 147–152. [Google Scholar] [CrossRef]
- Horsch, R.B.; Fraley, R.T.; Rogers, S.G.; Sanders, P.R.; Lloyd, A.; Hoffmann, N. Inheritance of functional foreign genes in plants. Science 1984, 223, 496–498. [Google Scholar] [CrossRef]
- Miedaner, T. Breeding strategies for improving plant resistance to diseases. In Advances in Plant Breeding Strategies: Agronomic, Abiotic and Biotic Stress Traits; Al-Khayri, J.M., Jain, S.M., Johnson, D.V., Eds.; Springer: Cham, Switzerland, 2016; pp. 561–599. [Google Scholar]
- Toriyama, K.; Arimoto, Y.; Uchimiya, H.; Hinata, K. Transgenic rice plants after direct gene transfer into protoplasts. Nat. Biotechnol. 1988, 6, 1072. [Google Scholar] [CrossRef]
- Gordon-Kamm, W.J.; Spencer, T.M.; Mangano, M.L.; Adams, T.R.; Daines, R.J.; Start, W.G.; O’Brien, J.V.; Chambers, S.A.; Adams, W.R.; Willetts, N.G. Transformation of maize cells and regeneration of fertile transgenic plants. Plant Cell 1990, 2, 603–618. [Google Scholar] [CrossRef] [PubMed]
- Vasilets, L.A.; Schwarz, W. Regulation of endogenous and expressed Na+/K+ pumps in Xenopus oocytes by membrane potential and stimulation of protein kinases. J. Membrane Biol. 1992, 125, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Rai, M.K.; Shekhawat, N. Recent advances in genetic engineering for improvement of fruit crops. Plant Cell Tiss. Org. 2014, 116, 1–15. [Google Scholar] [CrossRef]
- Peña, L.; Martín-Trillo, M.; Juárez, J.; Pina, J.A.; Navarro, L.; Martínez-Zapater, J.M. Constitutive expression of Arabidopsis LEAFY or APETALA1 genes in citrus reduces their generation time. Nat. Biotechnol. 2001, 19, 263. [Google Scholar] [CrossRef] [PubMed]
- Alquezar, B.; Rodrigo, M.J.; Zacarías, L. Regulation of carotenoid biosynthesis during fruit maturation in the red-fleshed orange mutant Cara Cara. Phytochemistry 2008, 69, 1997–2007. [Google Scholar] [CrossRef] [PubMed]
- Pons, E.; Alquézar, B.; Rodríguez, A.; Martorell, P.; Genovés, S.; Ramón, D.; Rodrigo, M.J.; Zacarías, L.; Pena, L. Metabolic engineering of β-carotene in orange fruit increases its in vivo antioxidant properties. Plant Biotechnol. J. 2014, 12, 17–27. [Google Scholar] [CrossRef]
- Dutt, M.; Barthe, G.; Irey, M.; Grosser, J. Transgenic Citrus Expressing an ArabidopsisNPR1 Gene Exhibit Enhanced Resistance against Huanglongbing (HLB; Citrus Greening). PLoS ONE 2015, 10, e0137134. [Google Scholar] [CrossRef]
- Alquézar, B.; Rodríguez, A.; de la Peña, M.; Peña, L. Genomic Analysis of Terpene Synthase Family and Functional Characterization of Seven Sesquiterpene Synthases from Citrus sinensis. Front. Plant Sci. 2017, 8, 1481. [Google Scholar] [CrossRef]
- Abel, P.P.; Nelson, R.S.; De, B.; Hoffmann, N.; Rogers, S.G.; Fraley, R.T.; Beachy, R.N. Delay of disease development in transgenic plants that express the tobacco mosaic virus coat protein gene. Science 1986, 232, 738–743. [Google Scholar] [CrossRef]
- Kobayashi, S.; Uchimiya, H. Expression and integration of a foreign gene in orange (Citrus sinensis Osb.) protoplasts by direct DNA transfer. Jpn. J. Genet. 1989, 64, 91–97. [Google Scholar] [CrossRef]
- Vardi, A.; Bleichman, S.; Aviv, D. Genetic transformation of Citrus protoplasts and regeneration of transgenic plants. Plant Sci. 1990, 69, 199–206. [Google Scholar] [CrossRef]
- Hidaka, T.; Omura, M.; Ugaki, M.; Tomiyama, M.; Kato, A.; Ohsima, M.; Motoyoshi, F. Agrobacterium-mediated transformation and regeneration of Citrus spp. from suspension cells. Jpn. J. Breed. 1990, 40, 199–207. [Google Scholar] [CrossRef]
- Hidaka, T.; Omura, M. Transformation of Citrus protoplasts by electroporation. J. Jpn. Soc. Hortic. Sci. 1993, 62, 371–376. [Google Scholar] [CrossRef]
- Yao, J.L.; Wu, J.H.; Gleave, A.P.; Morris, B.A. Transformation of citrus embryogenic cells using particle bombardment and production of transgenic embryos. Plant Sci. 1996, 113, 175–183. [Google Scholar] [CrossRef]
- Almeida, W.A.B.D.; Mourão Filho, F.D.A.A.; Mendes, B.M.J.; Pavan, A.; Rodriguez, A.P.M. Agrobacterium-mediated transformation of Citrus sinensis and Citrus limonia epicotyl segments. Sci. Agr. 2003, 60, 23–29. [Google Scholar] [CrossRef]
- Molinari, H.; Bespalhok, J.; Kobayashi, A.; Pereira, L.; Vieira, L. Agrobacteriumtumefaciens-mediated transformation of Swingle citrumelo (Citrus paradisi Macf. × Poncirustrifoliata L. Raf.) using thin epicotyl sections. Sci. Hortic. Amst. 2004, 99, 379–385. [Google Scholar] [CrossRef]
- Ahmad, M.; Mirza, B. An efficient protocol for transient transformation of intact fruit and transgene expression in Citrus. Plant Mol. Biol. Rep. 2005, 23, 419–420. [Google Scholar] [CrossRef]
- Dutt, M.; Grosser, J. An embryogenic suspension cell culture system for Agrobacterium-mediated transformation of citrus. Plant Cell Rep. 2010, 29, 1251–1260. [Google Scholar] [CrossRef]
- Al Bachchu, M.A.; Jin, S.B.; Park, J.W.; Sun, H.J.; Yun, S.H.; Lee, H.Y.; Lee, D.S.; Hong, Q.C.; Kim, Y.W.; Riu, K.Z. Agrobacterium-mediated transformation using embryogenic calli in satsuma mandarin (Citrus unshiu Marc.) cv. Miyagawa Wase. Hortic. Environ. Biotechnol. 2011, 52, 170–175. [Google Scholar] [CrossRef]
- He, Y.; Chen, S.; Peng, A.; Zou, X.; Xu, L.; Lei, T.; Liu, X.; Yao, L. Production and evaluation of transgenic sweet orange (Citrus sinensis Osbeck) containing bivalent antibacterial peptide genes (Shiva A and Cecropin B) via a novel Agrobacterium-mediated transformation of mature axillary buds. Sci. Hortic. Amst. 2011, 128, 99–107. [Google Scholar] [CrossRef]
- Fávero, P.; Mourão Filho, F.D.A.A.; Stipp, L.C.L.; Mendes, B.M.J. Genetic transformation of three sweet orange cultivars from explants of adult plants. Acta. Physiol. Plant. 2012, 34, 471–477. [Google Scholar] [CrossRef]
- Khan, E.U.; Fu, X.Z.; Liu, J.H. Agrobacterium-mediated genetic transformation and regeneration of transgenic plants using leaf segments as explants in Valencia sweet orange. Plant Cell Tiss. Org. 2012, 109, 383–390. [Google Scholar] [CrossRef]
- Lopez, C.; Cervera, M.; Fagoaga, C.; Moreno, P.; Navarro, L.; Flores, R.; Pena, L. Accumulation of transgene-derived siRNAs is not sufficient for RNAi-mediated protection against Citrus tristeza virus in transgenic Mexican lime. Mol. Plant Pathol. 2010, 11, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Soler, N.; Plomer, M.; Fagoaga, C.; Moreno, P.; Navarro, L.; Flores, R.; Peña, L. Transformation of Mexican lime with an intron-hairpin construct expressing untranslatable versions of the genes coding for the three silencing suppressors of Citrus tristeza virus confers complete resistance to the virus. Plant Biotechnol. J. 2012, 10, 597–608. [Google Scholar] [CrossRef] [PubMed]
- Donmez, D.; Simsek, O.; Izgu, T.; Aka Kacar, Y.; Yalcin Mendi, Y. Genetic transformation in citrus. Sci. World J. 2013, 2013, 491207. [Google Scholar] [CrossRef]
- Arora, L.; Narula, A. Gene editing and crop improvement using CRISPR-Cas9 system. Front. Plant Sci. 2017, 8, 1932. [Google Scholar] [CrossRef]
- Limera, C.; Sabbadini, S.; Sweet, J.B.; Mezzetti, B. New biotechnological tools for the genetic improvement of major woody fruit species. Front. Plant Sci. 2017, 8, 1418. [Google Scholar] [CrossRef]
- Yin, K.; Gao, C.; Qiu, J.L. Progress and prospects in plant genome editing. Nat. Plants 2017, 3, 17107. [Google Scholar] [CrossRef]
- Jia, H.; Wang, N. Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS ONE 2014, 9, e93806. [Google Scholar] [CrossRef]
- Jia, H.; Orbovic, V.; Jones, J.B.; Wang, N. Modification of the PthA4 effector binding elements in Type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccΔpthA4:dCs LOB1.3 infection. Plant Biotechnol. J. 2016, 14, 1291–1301. [Google Scholar] [CrossRef]
- Jia, H.; Zhang, Y.; Orbovic, V.; Xu, J.; White, F.F.; Jones, J.B.; Wang, N. Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 2017, 15, 817–823. [Google Scholar] [CrossRef] [PubMed]
- Peng, A.; Chen, S.; Lei, T.; Xu, L.; He, Y.; Wu, L.; Yao, L.; Zou, X. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 2017, 15, 1509–1519. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Orbović, V.; Wang, N. CRISPR-LbCas12a-mediated modification of citrus. Plant Biotechnol. J. 2019. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, S.; Peng, A.; Xie, Z.; He, Y.; Zou, X. CRISPR/Cas9-mediated editing of CsWRKY22 reduces susceptibility to Xanthomonas citri subsp. citri in Wanjincheng orange (Citrus sinensis (L.) Osbeck). Plant Biotechnol. Rep. 2019, 1–10. [Google Scholar] [CrossRef]
- Gottwald, T.R.; Graham, J.H.; Schubert, T.S. Citrus canker: The pathogen and its impact. Plant Health Progress 2002, 10, 32. [Google Scholar] [CrossRef]
- Das, A.K. Citrus canker-A review. J. Appl. Hort. 2003, 5, 52–60. [Google Scholar]
- Dutta, S.; Bendre, R.; Padhye, S.; Ahmed, F.; Sarkar, F. Synthesis, antioxidant properties and antiproliferative activities of tetrameric copper and copper-zinc metal complexes of catecholamine Schiff base ligand. Synth. React. Inorg. M. 2005, 35, 3–10. [Google Scholar] [CrossRef]
- Rinaldi, D.; Leite, R., Jr. Adaptation of Xanthomonas axonopodis pv. citri population to the presence of copper compounds in nature. Proc. Int. Soc. Citric 2000, 2, 1064. [Google Scholar]
- Behlau, F.; Belasque, J., Jr.; Bergamin Filho, A.; Graham, J.; Leite, R., Jr.; Gottwald, T. Copper sprays and windbreaks for control of citrus canker on young orange trees in southern Brazil. Crop Prot. 2008, 27, 807–813. [Google Scholar] [CrossRef]
- Zhang, X.; Francis, M.I.; Dawson, W.O.; Graham, J.H.; Orbović, V.; Triplett, E.W.; Mou, Z. Over-expression of the Arabidopsis NPR1 gene in citrus increases resistance to citrus canker. Eur. J. Plant Pathol. 2010, 128, 91–100. [Google Scholar] [CrossRef]
- Zasloff, M. Antibiotic peptides as mediators of innate immunity. Curr. Opin. Immunol. 1992, 4, 3–7. [Google Scholar] [CrossRef]
- Boman, H. Antibacterial peptides: Basic facts and emerging concepts. J. Intern. Med. 2003, 254, 197–215. [Google Scholar] [CrossRef] [PubMed]
- Boscariol, R.L.; Monteiro, M.; Takahashi, E.K.; Chabregas, S.M.; Vieira, M.L.C.; Vieira, L.G.E.; Pereira, L.F.P.; Mourao Filho, F.D.A.A.; Cardoso, S.C.; Christiano, R.S.C. Attacin A gene from Tricloplusia ni reduces susceptibility to Xanthomonas axonopodis pv. citri in transgenic Citrus sinensis ‘Hamlin’. J. Am. Soc. Hortic. Sci. 2006, 131, 530–536. [Google Scholar] [CrossRef]
- Cardoso, S.C.; Barbosa-Mendes, J.M.; Boscariol-Camargo, R.L.; Christiano, R.S.C.; Filho, A.B.; Vieira, M.L.C.; Mendes, B.M.J.; Mourão Filho, F.D.A.A. Transgenic Sweet Orange (Citrus sinensis L. Osbeck) Expressing the attacin A Gene for Resistance to Xanthomonas citri subsp. citri. Plant Mol. Biol. Rep. 2009, 28, 185–192. [Google Scholar] [CrossRef]
- Sala Junior, V.; Celloto, V.R.; Vieira, L.G.E.; Gonçalves, J.E.; Gonçalves, R.A.C.; de Oliveira, A.J.B. Floral nectar chemical composition of floral nectar in conventional and transgenic sweet orange, Citrus sinensis (L.) Osbeck, expressing an antibacterial peptide. Plant Syst. Evol. 2008, 275, 1–7. [Google Scholar] [CrossRef]
- Kobayashi, A.K.; Vieira, L.G.E.; Bespalhok Filho, J.C.; Leite, R.P.; Pereira, L.F.P.; Molinari, H.B.C.; Marques, V.V. Enhanced resistance to citrus canker in transgenic sweet orange expressing the sarcotoxin IA gene. Eur. J. Plant Pathol. 2017, 149, 865–873. [Google Scholar] [CrossRef] [Green Version]
- Hao, G.; Zhang, S.; Stover, E. Transgenic expression of antimicrobial peptide D2A21 confers resistance to diseases incited by Pseudomonas syringae pv. tabaci and Xanthomonas citri, but not Candidatus Liberibacter asiaticus. PLoS ONE 2017, 12, e0186810. [Google Scholar]
- Furman, N.; Kobayashi, K.; Zanek, M.C.; Calcagno, J.; Garcia, M.L.; Mentaberry, A. Transgenic sweet orange plants expressing a dermaseptin coding sequence show reduced symptoms of citrus canker disease. J. Biotechnol. 2013, 167, 412–419. [Google Scholar] [CrossRef]
- Chern, M.; Fitzgerald, H.A.; Canlas, P.E.; Navarre, D.A.; Ronald, P.C. Over expression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol. Plant Microbe Interact. 2005, 18, 511–520. [Google Scholar] [CrossRef]
- Lin, W.C.; Lu, C.F.; Wu, J.W.; Cheng, M.L.; Lin, Y.M.; Yang, N.S.; Black, L.; Green, S.K.; Wang, J.F.; Cheng, C.P. Transgenic tomato plants expressing the Arabidopsis NPR1 gene display enhanced resistance to a spectrum of fungal and bacterial diseases. Transgenic Res. 2004, 13, 567–581. [Google Scholar] [CrossRef]
- Makandar, R.; Essig, J.S.; Schapaugh, M.A.; Trick, H.N.; Shah, J. Genetically engineered resistance to Fusarium head blight in wheat by expression of Arabidopsis NPR1. Mol. Plant Microbe Interact. 2006, 19, 123–129. [Google Scholar] [CrossRef] [PubMed]
- Malnoy, M.; Jin, Q.; Borejsza-Wysocka, E.; He, S.; Aldwinckle, H. Over expression of the apple MpNPR1 gene confers increased disease resistance in Malus × domestica. Mol. Plant Microbe Interact. 2007, 20, 1568–1580. [Google Scholar] [CrossRef] [PubMed]
- Potlakayala, S.D.; DeLong, C.; Sharpe, A.; Fobert, P.R. Conservation of NON-EXPRESSOR OF PATHOGENESIS-RELATED GENES1 function between Arabidopsis thaliana and Brassica napus. Physiol. Mol. Plant Pathol. 2007, 71, 174–183. [Google Scholar] [CrossRef]
- Chen, X.; Barnaby, J.Y.; Sreedharan, A.; Huang, X.; Orbović, V.; Grosser, J.W.; Wang, N.; Dong, X.; Song, W.Y. Over-expression of the citrus gene CtNH1 confers resistance to bacterial canker disease. Physiol. Mol. Plant Pathol. 2013, 84, 115–122. [Google Scholar] [CrossRef]
- Flor, H.H. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 1971, 9, 275–296. [Google Scholar] [CrossRef]
- Tai, T.H.; Dahlbeck, D.; Clark, E.T.; Gajiwala, P.; Pasion, R.; Whalen, M.C.; Stall, R.E.; Staskawicz, B.J. Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc. Natl. Acad. Sci. USA 1999, 96, 14153–14158. [Google Scholar] [CrossRef]
- Sendín, L.; Filippone, M.; Orce, I.; Rigano, L.; Enrique, R.; Peña, L.; Vojnov, A.; Marano, M.; Castagnaro, A. Transient expression of pepper Bs2 gene in Citrus limon as an approach to evaluate its utility for management of citrus canker disease. Plant Pathol. 2012, 61, 648–657. [Google Scholar] [CrossRef]
- Sendín, L.N.; Orce, I.G.; Gómez, R.L.; Enrique, R.; Bournonville, C.F.G.; Noguera, A.S.; Vojnov, A.A.; Marano, M.R.; Castagnaro, A.P.; Filippone, M.P. Inducible expression of Bs2 R gene from Capsicum chacoense in sweet orange (Citrus sinensis L. Osbeck) confers enhanced resistance to citrus canker disease. Plant Mol. Biol. 2017, 93, 607–621. [Google Scholar] [CrossRef]
- Song, W.Y.; Wang, G.L.; Chen, L.L.; Kim, H.S.; Pi, L.Y.; Holsten, T.; Gardner, J.; Wang, B.; Zhai, W.X.; Zhu, L.H. A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 1995, 270, 1804–1806. [Google Scholar] [CrossRef]
- Omar, A.; Song, W.Y.; Grosser, J. Introduction of Xa21, a Xanthomonas-resistance gene from rice, into ‘Hamlin’sweet orange [Citrus sinensis (L.) Osbeck] using protoplast-GFP co-transformation or single plasmid transformation. J. Hortic. Sci. Biotech. 2007, 82, 914–923. [Google Scholar] [CrossRef]
- Mendes, B.M.J.; Cardoso, S.; Boscariol-Camargo, R.; Cruz, R.; Mourão Filho, F.; Bergamin Filho, A. Reduction in susceptibility to Xanthomonas axonopodis pv. citri in transgenic Citrus sinensis expressing the rice Xa21 gene. Plant Pathol. 2010, 59, 68–75. [Google Scholar]
- Li, D.L.; Xiao, X.; Guo, W.W. Production of Transgenic Anliucheng Sweet Orange (Citrus sinensis Osbeck) with Xa21 Gene for Potential Canker Resistance. J. Integr. Agr. 2014, 13, 2370–2377. [Google Scholar] [CrossRef]
- Omar, A.A.; Murata, M.M.; El-Shamy, H.A.; Graham, J.H.; Grosser, J.W. Enhanced resistance to citrus canker in transgenic mandarin expressing Xa21 from rice. Transgenic Res. 2018, 27, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Apel, K.; Hirt, H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 2004, 55, 373–399. [Google Scholar] [CrossRef]
- Gechev, T.S.; Van Breusegem, F.; Stone, J.M.; Denev, I.; Laloi, C. Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays 2006, 28, 1091–1101. [Google Scholar] [CrossRef]
- Hao, G.; Pitino, M.; Duan, Y.; Stover, E. Reduced susceptibility to Xanthomonas citri in transgenic citrus expressing the FLS2 receptor from Nicotiana benthamiana. Mol. Plant Microbe Interact. 2016, 29, 132–142. [Google Scholar] [CrossRef]
- De Oliveira, M.L.; de Lima Silva, C.C.; Abe, V.Y.; Costa, M.G.; Cernadas, R.A.; Benedetti, C.E. Increased resistance against citrus canker mediated by a citrus mitogen-activated protein kinase. Mol. Plant Microbe Interact. 2013, 26, 1190–1199. [Google Scholar] [CrossRef]
- Barbosa-Mendes, J.M.; Filho, F.D.A.A.M.; Filho, A.B.; Harakava, R.; Beer, S.V.; Mendes, B.M.J. Genetic transformation of Citrus sinensis cv. Hamlin with hrpN gene from Erwinia amylovora and evaluation of the transgenic lines for resistance to citrus canker. Sci. Hortic. Amst. 2009, 122, 109–115. [Google Scholar] [CrossRef]
- Fu, X.Z.; Chen, C.W.; Wang, Y.; Liu, J.H.; Moriguchi, T. Ectopic expression of MdSPDS1 in sweet orange (Citrus sinensis Osbeck) reduces canker susceptibility: Involvement of H2O2 production and transcriptional alteration. BMC Plant Biol. 2011, 11, 55. [Google Scholar] [CrossRef]
- Qin, L.; Luo, S.; Deng, Z.; Yan, J.; Li, N.; Yuan, F. Enhanced resistance to canker disease and tolerances to biotic and abiotic stresses in terf1 transgenic sweet orange. In II International Symposium on Citrus Biotechnology; Gentile, A., La Malfa, S., Eds.; International Society for Horticultural Science: Catania, Italy, 2009; pp. 165–172. [Google Scholar]
- Yang, L.; Hu, C.; Li, N.; Zhang, J.; Yan, J.; Deng, Z. Transformation of sweet orange [Citrus sinensis (L.) Osbeck] with pthA-nls for acquiring resistance to citrus canker disease. Plant Mol. Biol. 2011, 75, 11–23. [Google Scholar] [CrossRef]
- Hao, G.; Stover, E.; Gupta, G. Overexpression of a modified plant thionin enhances disease Resistance to citrus canker and huanglongbing (HLB). Front. Plant Sci. 2016, 7, 1078. [Google Scholar] [CrossRef] [PubMed]
- Bové, J.M. Huanglongbing: A destructive, newly-emerging, century-old disease of citrus. J. Plant Pathol. 2006, 88, 7–37. [Google Scholar]
- Robertson, C.J.; Zhang, X.; Gowda, S.; Orbović, V.; Dawson, W.O.; Mou, Z. Overexpression of the Arabidopsis NPR1 protein in citrus confers tolerance to Huanglongbing. J. Citrus Pathol. 2018, 5, 1–8. [Google Scholar]
- Munir, S.; He, P.; Wu, Y.; He, P.; Khan, S.; Huang, M.; Cui, W.; He, P.; He, Y. Huanglongbing control: Perhaps the end of the beginning. Microb. Ecol. 2018, 76, 192–204. [Google Scholar] [CrossRef]
- Gottwald, T.R. Current epidemiological understanding of citrus huanglongbing. Annu. Rev. Phytopathol. 2010, 48, 119–139. [Google Scholar] [CrossRef]
- Wang, N.; Trivedi, P. Citrus huanglongbing: A newly relevant disease presents unprecedented challenges. Phytopathology 2013, 103, 652–665. [Google Scholar] [CrossRef]
- Da Graça, J.; Korsten, L. Citrus huanglongbing: Review, present status and future strategies. In Diseases of Fruits and Vegetables Volume I; Naqvi, S.A.M.H., Ed.; Springer: Dordrecht, The Netherlands, 2004; pp. 229–245. [Google Scholar]
- Zhang, M.; Guo, Y.; Powell, C.A.; Doud, M.S.; Yang, C.; Duan, Y. Effective antibiotics against ‘Candidatus Liberibacter asiaticus’ in HLB-affected citrus plants identified via the graft-based evaluation. PLoS ONE 2014, 9, e111032. [Google Scholar] [CrossRef]
- Miles, G.P.; Stover, E.; Ramadugu, C.; Keremane, M.L.; Lee, R.F. Apparent tolerance to Huanglongbing in Citrus and Citrus-related germplasm. HortScience 2017, 52, 31–39. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, L.; Yu, X.; Stover, E.; Luo, F.; Duan, Y. Transcriptome profiling of Huanglongbing (HLB) tolerant and susceptible citrus plants reveals the role of basal resistance in HLB tolerance. Front. Plant Sci. 2016, 7, 933. [Google Scholar] [CrossRef]
- Felipe, R.T.A.; Mourão Filho, F.D.A.A.; Lopes, S.A.; Mendes, B.M.J.; Behling, M.; Pereira Junior, E.V. Reaction of sweet orange cultivars expressing the attacin A gene to ‘Candidatus Liberibacter asiaticus’ infection. Pesqui. Agropecu. Bras. 2013, 48, 1440–1448. [Google Scholar] [CrossRef]
- Rocha Tavano, E.C.D.; Erpen, L.; Aluisi, B.; Harakava, R.; Lopes, J.R.S.; Vieira, M.L.C.; Piedade, S.M.D.S.; Mendes, B.M.J.; Mourao Filho, A.A. Sweet orange genetic transformation with the attacin A gene under the control of phloem-specific promoters and inoculation with Candidatus Liberibacter asiaticus. J. Hortic. Sci. Biotech. 2019, 94, 210–219. [Google Scholar] [CrossRef]
- Zou, X.; Jiang, X.; Xu, L.; Lei, T.; Peng, A.; He, Y.; Yao, L.; Chen, S. Transgenic citrus expressing synthesized cecropin B genes in the phloem exhibits decreased susceptibility to Huanglongbing. Plant Mol. Biol. 2017, 93, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Bar-Joseph, M.; Marcus, R.; Lee, R.F. The continuous challenge of citrus tristeza virus control. Annu. Rev. Phytopathol. 1989, 27, 291–316. [Google Scholar] [CrossRef]
- Dominguez, A.; Guerri, J.; Cambra, M.; Navarro, L.; Moreno, P.; Pena, L. Efficient production of transgenic citrus plants expressing the coat protein gene of citrus tristeza virus. Plant Cell Rep. 2000, 19, 427–433. [Google Scholar] [CrossRef]
- Domínguez, A.; de Mendoza, A.H.; Guerri, J.; Cambra, M.; Navarro, L.; Moreno, P.; Peña, L. Pathogen-derived resistance to Citrus tristeza virus (CTV) in transgenic Mexican lime (Citrus aurantifolia (Christ.) Swing.) plants expressing its p25 coat protein gene. Mol. Breed. 2002, 10, 1–10. [Google Scholar]
- Fagoaga, C.; López, C.; de Mendoza, A.H.; Moreno, P.; Navarro, L.; Flores, R.; Peña, L. Post-transcriptional gene silencing of the p23 silencing suppressor of Citrus tristeza virus confers resistance to the virus in transgenic Mexican lime. Plant Mol. Biol. 2006, 60, 153–165. [Google Scholar] [CrossRef]
- Ananthakrishnan, G.; Orbović, V.; Pasquali, G.; Ćalović, M.; Grosser, J. Transfer of citrus tristeza virus (CTV)-derived resistance candidate sequences to four grapefruit cultivars through Agrobacterium-mediated genetic transformation. In Vitro Cell. Dev. Plant 2007, 43, 593–601. [Google Scholar] [CrossRef]
- Muniz, F.; De Souza, A.; Stipp, L.C.L.; Schinor, E.; Freitas, W.; Harakava, R.; Stach-Machado, D.; Rezende, J.A.M.; Mourão Filho, F.; Mendes, B. Genetic transformation of Citrus sinensis with Citrus tristeza virus (CTV) derived sequences and reaction of transgenic lines to CTV infection. Biol. Plant. 2012, 56, 162–166. [Google Scholar] [CrossRef]
- Olivares-Fuster, O.; Fleming, G.; Albiach-Marti, M.; Gowda, S.; Dawson, W.; Crosser, J. Citrus tristeza virus (CTV) resistance in transgenic citrus based on virus challenge of protoplasts. In Vitro Cell. Dev. Plant 2003, 39, 567–572. [Google Scholar] [CrossRef]
- Cervera, M.; Esteban, O.; Gil, M.; Gorris, M.T.; Martínez, M.C.; Peña, L.; Cambra, M. Transgenic expression in citrus of single-chain antibody fragments specific to Citrus tristeza virus confers virus resistance. Transgenic Res. 2010, 19, 1001–1015. [Google Scholar] [CrossRef]
- Waterhouse, P.M.; Wang, M.B.; Lough, T. Gene silencing as an adaptive defence against viruses. Nature 2001, 411, 834. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Zhang, Y.; Yang, J.; Zhong, Y. Expression of hairpin RNA (hpRNA) targeting the three CTV-silencing suppressor genes confers sweet orange with stem-pitting CTV tolerance. J. Hortic. Sci. Biotechnol. 2017, 92, 465–474. [Google Scholar] [CrossRef]
- Cheng, C.Z.; Yang, J.W.; Yan, H.B.; Bei, X.J.; Zhang, Y.Y.; Lu, Z.M.; Zhong, G.Y. Expressing p20 hairpin RNA of Citrus tristeza virus confers Citrus aurantium with tolerance/resistance against stem pitting and seedling yellow CTV strains. J. Integr. Agric. 2015, 14, 1767–1777. [Google Scholar] [CrossRef]
- Kayim, M.; Barthe, G.; Beretta, J.; Derrick, K. Transgenic citrus plants expressing the coat protein gene of Citrus psorosis virus. Phytopathology 2005, 95, S52. [Google Scholar]
- Reyes, C.A.; Zanek, M.C.; Velázquez, K.; Costa, N.; Plata, M.I.; Garcia, M.L. Generation of sweet orange transgenic lines and evaluation of citrus psorosis virus-derived resistance against psorosis A and psorosis B. J. Phytopathol. 2011, 159, 531–537. [Google Scholar] [CrossRef]
- Reyes, C.A.; De Francesco, A.; Peña, E.J.; Costa, N.; Plata, M.I.; Sendin, L.; Castagnaro, A.P.; García, M.L. Resistance to Citrus psorosis virus in transgenic sweet orange plants is triggered by coat protein–RNA silencing. J. Biotechnol. 2011, 151, 151–158. [Google Scholar] [CrossRef]
- Gentile, A.; Deng, Z.; La Malfa, S.; Distefano, G.; Domina, F.; Vitale, A.; Polizzi, G.; Lorito, M.; Tribulato, E. Enhanced resistance to Phoma tracheiphila and Botrytis cinerea in transgenic lemon plants expressing a Trichoderma harzianum chitinase gene. Plant Breed. 2007, 126, 146–151. [Google Scholar] [CrossRef]
- Distefano, G.; La Malfa, S.; Vitale, A.; Lorito, M.; Deng, Z.; Gentile, A. Defence-related gene expression in transgenic lemon plants producing an antimicrobial Trichoderma harzianum endochitinase during fungal infection. Transgenic Res. 2008, 17, 873–879. [Google Scholar] [CrossRef]
- Fagoaga, C.; Rodrigo, I.; Conejero, V.; Hinarejos, C.; Tuset, J.J.; Arnau, J.; Pina, J.A.; Navarro, L.; Peña, L. Increased tolerance to Phytophthora citrophthora in transgenic orange plants constitutively expressing a tomato pathogenesis related protein PR-5. Mol. Breed. 2001, 7, 175–185. [Google Scholar] [CrossRef]
- Mondal, S.; Dutt, M.; Grosser, J.; Dewdney, M. Transgenic citrus expressing the antimicrobial gene AttacinE (attE) reduces the susceptibility of ‘Duncan’ grapefruit to the citrus scab caused by Elsinoë fawcettii. Eur. J. Plant Pathol. 2012, 133, 391–404. [Google Scholar] [CrossRef]
- Rodríguez, A.; Andrés, V.S.; Cervera, M.; Redondo, A.; Alquézar, B.; Shimada, T.; Gadea, J.; Rodrigo, M.; Zacarías, L.; Palou, L. The monoterpene limonene in orange peels attracts pests and microorganisms. Plant Signal. Behav. 2011, 6, 1820–1823. [Google Scholar] [CrossRef] [Green Version]
- Parmar, N.; Singh, K.H.; Sharma, D.; Singh, L.; Kumar, P.; Nanjundan, J.; Khan, Y.J.; Chauhan, D.K.; Thakur, A.K. Genetic engineering strategies for biotic and abiotic stress tolerance and quality enhancement in horticultural crops: A comprehensive review. 3 Biotech 2017, 7, 239. [Google Scholar] [CrossRef]
- Jansing, J.; Schiermeyer, A.; Schillberg, S.; Fischer, R.; Bortesi, L. Genome Editing in Agriculture: Technical and Practical Considerations. Int. J. Mol. Sci. 2019, 20, 2888. [Google Scholar] [CrossRef]
- Pennisi, E. The CRISPR Craze. Science 2013, 341, 833–836. [Google Scholar] [CrossRef]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013, 41, e188. [Google Scholar] [CrossRef]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014, 32, 947. [Google Scholar] [CrossRef]
- Char, S.N.; Neelakandan, A.K.; Nahampun, H.; Frame, B.; Main, M.; Spalding, M.H.; Becraft, P.W.; Meyers, B.C.; Walbot, V.; Wang, K. An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnol. J. 2017, 15, 257–268. [Google Scholar] [CrossRef]
- Nishitani, C.; Hirai, N.; Komori, S.; Wada, M.; Okada, K.; Osakabe, K.; Yamamoto, T.; Osakabe, Y. Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 2016, 6, 31481. [Google Scholar] [CrossRef]
- Ren, C.; Liu, X.; Zhang, Z.; Wang, Y.; Duan, W.; Li, S.; Liang, Z. CRISPR/Cas9-mediated efficient targeted mutagenesis in Chardonnay (Vitis vinifera L.). Sci. Rep. 2016, 6, 32289. [Google Scholar] [CrossRef]
- Song, G.; Jia, M.; Chen, K.; Kong, X.; Khattak, B.; Xie, C.; Li, A.; Mao, L. CRISPR/Cas9: A powerful tool for crop genome editing. Crop J. 2016, 4, 75–82. [Google Scholar] [CrossRef] [Green Version]
- Jia, H.; Zou, X.; Orbovic, V.; Wang, N. Genome Editing in Citrus Tree with CRISPR/Cas9. In Plant Genome Editing with CRISPR Systems; Qi, Y., Ed.; Methods in Molecular Biology; Humana Press: New York, NY, USA, 2019; Volume 1917, pp. 235–241. [Google Scholar]
- Xu, Z.S.; Feng, K.; Xiong, A.S. CRISPR/Cas9-mediated multiply targeted mutagenesis in orange and purple carrot plants. Mol. Biotechnol. 2019, 61, 191–199. [Google Scholar] [CrossRef]
- Knott, G.J.; Doudna, J.A.J.S. CRISPR-Cas guides the future of genetic engineering. Science 2018, 361, 866–869. [Google Scholar] [CrossRef] [Green Version]
- Ali, Z.; Abulfaraj, A.; Idris, A.; Ali, S.; Tashkandi, M.; Mahfouz, M.M. CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 2015, 16, 238. [Google Scholar] [CrossRef] [Green Version]
- Baltes, N.J.; Hummel, A.W.; Konecna, E.; Cegan, R.; Bruns, A.N.; Bisaro, D.M.; Voytas, D.F. Conferring resistance to geminiviruses with the CRISPR–Cas prokaryotic immune system. Nat. Plants 2015, 1, 15145. [Google Scholar] [CrossRef]
- Malnoy, M.; Viola, R.; Jung, M.H.; Koo, O.J.; Kim, S.; Kim, J.S.; Velasco, R.; Nagamangala Kanchiswamy, C. DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 2016, 7, 1904. [Google Scholar] [CrossRef]
- Jia, H.; Wang, N. Xcc-facilitated agroinfiltration of citrus leaves: A tool for rapid functional analysis of transgenes in citrus leaves. Plant Cell Rep. 2014, 33, 1993–2001. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, J.; Jia, H.; Sosso, D.; Li, T.; Frommer, W.B.; Yang, B.; White, F.F.; Wang, N.; Jones, J.B. Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. USA 2014, 111, E521–E529. [Google Scholar] [CrossRef]
- Ledford, H. Geneticists enlist engineered virus and CRISPR to battle citrus disease. Nat. News 2017, 545, 277. [Google Scholar] [CrossRef]
- Wang, N.; Pierson, E.A.; Setubal, J.C.; Xu, J.; Levy, J.G.; Zhang, Y.; Li, J.; Rangel, L.T.; Martins, J., Jr. The Candidatus Liberibacter–host interface: Insights into pathogenesis mechanisms and disease control. Annu. Rev. Phytopathol. 2017, 55, 451–482. [Google Scholar] [CrossRef]
- Zhang, Y.; Malzahn, A.A.; Sretenovic, S.; Qi, Y. The emerging and uncultivated potential of CRISPR technology in plant science. Nat. Plants 2019, 5, 778–794. [Google Scholar] [CrossRef]
- Zhu, C.; Zheng, X.; Huang, Y.; Ye, J.; Chen, P.; Zhang, C.; Zhao, F.; Xie, Z.; Zhang, S.; Wang, N. Genome sequencing and CRISPR/Cas9 gene editing of an early flowering Mini-Citrus (Fortunella hindsii). Plant Biotechnol. J. 2019, 17, 2199–2210. [Google Scholar] [CrossRef]
- Hunter, W.B.; Sinisterra-Hunter, X.H. Emerging RNA Suppression Technologies to Protect Citrus Trees from Citrus Greening Disease Bacteria. Adv. Insect Physiol. 2018, 55, 163–197. [Google Scholar]
- Goulin, E.H.; Galdeano, D.M.; Granato, L.M.; Matsumura, E.E.; Dalio, R.J.D.; Machado, M.A. RNA interference and CRISPR: Promising approaches to better understand and control citrus pathogens. Microbiol. Res. 2019, 226, 1–9. [Google Scholar] [CrossRef]
- Andrade, C.; Hunter, W.B. RNA interference-natural gene-based technology for highly specific pest control (HiSPeC). RNA Interf. 2016, 391–409. [Google Scholar] [CrossRef]
- Andrade, E.C.; Hunter, W.B. RNAi feeding bioassay: Development of a non-transgenic approach to control Asian citrus psyllid and other hemipterans. Entomol. Exp. Appl. 2017, 162, 389–396. [Google Scholar] [CrossRef]
- Galdeano, D.M.; Breton, M.C.; Lopes, J.R.S.; Falk, B.W.; Machado, M.A. Oral delivery of double-stranded RNAs induces mortality in nymphs and adults of the Asian citrus psyllid, Diaphorina citri. PLoS ONE 2017, 12, e0171847. [Google Scholar] [CrossRef]
- Ghosh, S.K.B.; Hunter, W.B.; Park, A.L.; Gundersen-Rindal, D.E. Double strand RNA delivery system for plant-sap-feeding insects. PLoS ONE 2017, 12, e0171861. [Google Scholar] [CrossRef]
- Ghosh, S.K.B.; Hunter, W.B.; Park, A.L.; Gundersen-Rindal, D.E. Double-stranded RNA oral delivery methods to induce RNA interference in phloem and plant-sap-feeding hemipteran insects. JoVE J. Vis. Exp. 2018, 135, e57390. [Google Scholar] [CrossRef]
- Kruse, A.; Fattah-Hosseini, S.; Saha, S.; Johnson, R.; Warwick, E.; Sturgeon, K.; Mueller, L.; MacCoss, M.J.; Shatters Jr, R.G.; Heck, M.C. Combining’omics and microscopy to visualize interactions between the Asian citrus psyllid vector and the Huanglongbing pathogen Candidatus Liberibacter asiaticus in the insect gut. PLoS ONE 2017, 12, e0179531. [Google Scholar] [CrossRef]
- Taning, C.N.; Andrade, E.C.; Hunter, W.B.; Christiaens, O.; Smagghe, G. Asian Citrus Psyllid RNAi Pathway-RNAi evidence. Sci. Rep. 2016, 6, 38082. [Google Scholar] [CrossRef]
- Ammar, E.D.; Walter, A.J.; Hall, D.G. New excised-leaf assay method to test inoculativity of Asian citrus psyllid (Hemiptera: Psyllidae) with Candidatus Liberibacter asiaticus associated with citrus huanglongbing disease. J. Econ. Entomol. 2013, 106, 25–35. [Google Scholar] [CrossRef]
- Raiol-Junior, L.L.; Baia, A.D.; Luiz, F.Q.; Fassini, C.G.; Marques, V.V.; Lopes, S.A. Improvement in the Excised Citrus Leaf Assay to Investigate Inoculation of ‘Candidatus Liberibacter asiaticus’ by the Asian Citrus Psyllid Diaphorina citri. Plant Dis. 2017, 101, 409–413. [Google Scholar] [CrossRef]
- Russell, C.W.; Pelz-Stelinski, K.S. Development of an artificial diet and feeding system for juvenile stages of the A sian citrus psyllid, Diaphorina citri. Entomol. Exp. Appl. 2015, 154, 171–176. [Google Scholar] [CrossRef]
- Killiny, N.; Hajeri, S.; Tiwari, S.; Gowda, S.; Stelinski, L.L. Double-stranded RNA uptake through topical application, mediates silencing of five CYP4 genes and suppresses insecticide resistance in Diaphorina citri. PLoS ONE 2014, 9, e110536. [Google Scholar] [CrossRef]
- Killiny, N.; Kishk, A. Delivery of dsRNA through topical feeding for RNA interference in the citrus sap piercing-sucking hemipteran, Diaphorina citri. Arch. Insect Biochem. 2017, 95, e21394. [Google Scholar] [CrossRef]
- Yu, X.; Gowda, S.; Killiny, N. Double-stranded RNA delivery through soaking mediates silencing of the muscle protein 20 and increases mortality to the Asian citrus psyllid, Diaphorina citri. Pest Manag. Sci. 2017, 73, 1846–1853. [Google Scholar] [CrossRef]
- Zhang, L.; Reed, R.D. A Practical Guide to CRISPR/Cas9 Genome Editing in Lepidoptera. In Diversity and Evolution of Butterfly Wing Patterns; Sekimura, T., Nijhout, H., Eds.; Springer: Singapore, 2017; pp. 155–172. [Google Scholar]
- Zheng, Z.; Bao, M.; Wu, F.; Chen, J.; Deng, X. Predominance of single prophage carrying a CRISPR/cas system in “Candidatus Liberibacter asiaticus” strains in southern China. PLoS ONE 2016, 11, e0146422. [Google Scholar] [CrossRef]
- Shang, F.; Xiong, Y.; Xia, W.K.; Wei, D.D.; Wei, D.; Wang, J.J. Identification, characterization and functional analysis of a chitin synthase gene in the brown citrus aphid, Toxoptera citricida (Hemiptera, Aphididae). Insect Mol. Biol. 2016, 25, 422–430. [Google Scholar] [CrossRef]
- Jing, T.X.; Tan, Y.; Ding, B.Y.; Dou, W.; Wei, D.; Wang, J.J. NADPH–cytochrome P450 reductase mediates the resistance of Aphis (Toxoptera) citricidus (Kirkaldy) to abamectin. Front. Plant Sci. 2018, 9, 986. [Google Scholar] [CrossRef]
- Rosa, C.; Kamita, S.G.; Dequine, H.; Wuriyanghan, H.; Lindbo, J.A.; Falk, B.W. RNAi effects on actin mRNAs in Homalodisca vitripennis cells. J. RNAi Gene Silenc. 2010, 6, 361. [Google Scholar]
- Rosa, C.; Kamita, S.G.; Falk, B.W. RNA interference is induced in the glassy winged sharpshooter Homalodisca vitripennis by actin dsRNA. Pest Manag. Sci. 2012, 68, 995–1002. [Google Scholar] [CrossRef]
- Roeschlin, R.A.; Uviedo, F.; García, L.; Molina, M.C.; Favaro, M.A.; Chiesa, M.A.; Tasselli, S.; Franco-Zorrilla, J.M.; Forment, J.; Gadea, J.; et al. PthA4AT, a 7.5-repeats transcription activator-like (TAL) effector from Xanthomonas citri ssp. citri, triggers citrus canker resistance. Mol. Plant Pathol. 2019, 20, 1394–1407. [Google Scholar] [CrossRef]
- Shimo, H.M.; Terassi, C.; Lima Silva, C.C.; de Lima Zanella, J.; Mercaldi, G.F.; Rocco, S.A.; Benedetti, C.E. Role of the Citrus sinensis RNA deadenylase CsCAF1 in citrus canker resistance. Mol. Plant Pathol. 2019, 20, 1105–1118. [Google Scholar] [CrossRef]
- Glandorf, D.; Bakker, P.; Van Loon, L.J. Influence of the production of antibacterial and antifungal proteins by transgenic plants on the saprophytic soil microflora. Acta Bot. Neerl. 1997, 46, 85–104. [Google Scholar] [CrossRef]
- Bulgarelli, D.J.S. How manipulating the plant microbiome could improve agriculture. Scientist 2018, 32, 51422. [Google Scholar]
- Ageitos, J.M.; Sanchez-Perez, A.; Calo-Mata, P.; Villa, T.G. Antimicrobial peptides (AMPs): Ancient compounds that represent novel weapons in the fight against bacteria. Biochem. Pharmacol. 2017, 133, 117–138. [Google Scholar] [CrossRef]
- Weinhold, A.; Dorcheh, E.K.; Li, R.; Rameshkumar, N.; Baldwin, I.T. Antimicrobial peptide expression in a wild tobacco plant reveals the limits of host-microbe-manipulations in the field. eLife 2018, 7, e28715. [Google Scholar] [CrossRef]
- Cunningham, F.J.; Goh, N.S.; Demirer, G.S.; Matos, J.L.; Landry, M.P. Nanoparticle-mediated delivery towards advancing plant genetic engineering. Trends Biotechnol. 2018, 36, 882–897. [Google Scholar] [CrossRef]
- Wang, J.W.; Grandio, E.G.; Newkirk, G.M.; Demirer, G.S.; Butrus, S.; Giraldo, J.P.; Landry, M.P. Nanoparticle-Mediated Genetic Engineering of Plants. Mol. Plant 2019, 12, 1037–1040. [Google Scholar] [CrossRef]
- Dyab, A.K.; Mohamed, L.A.; Taha, F. Non-aqueous olive oil-in-glycerin (o/o) Pickering emulsions: Preparation, characterization and in vitro aspirin release. J. Disper. Sci. Technol. 2018, 39, 890–900. [Google Scholar] [CrossRef]
- Naqvi, S.; Maitra, A.; Abdin, M.; Akmal, M.; Arora, I.; Samim, M. Calcium phosphate nanoparticle mediated genetic transformation in plants. J. Mater. Chem. 2012, 22, 3500–3507. [Google Scholar] [CrossRef]
- Pasupathy, K.; Lin, S.; Hu, Q.; Luo, H.; Ke, P.C. Direct plant gene delivery with a poly (amidoamine) dendrimer. Biotechnol. J. Healthc. Nutr. Technol. 2008, 3, 1078–1082. [Google Scholar] [CrossRef]
- Burlaka, O.; Pirko, Y.V.; Yemets, A.; Blume, Y.B. Plant genetic transformation using carbon nanotubes for DNA delivery. Cytol. Genet. 2015, 49, 349–357. [Google Scholar] [CrossRef]
- Chang, F.P.; Kuang, L.Y.; Huang, C.A.; Jane, W.N.; Hung, Y.; Yue-ie, C.H.; Mou, C.Y. A simple plant gene delivery system using mesoporous silica nanoparticles as carriers. J. Mater. Chem. B 2013, 1, 5279–5287. [Google Scholar] [CrossRef]
- Jiang, L.; Ding, L.; He, B.; Shen, J.; Xu, Z.; Yin, M.; Zhang, X. Systemic gene silencing in plants triggered by fluorescent nanoparticle-delivered double-stranded RNA. Nanoscale 2014, 6, 9965–9969. [Google Scholar] [CrossRef]
- Gao, Y.; Zhao, Y. Specific and heritable gene editing in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 4357–4358. [Google Scholar] [CrossRef]
- Kanchiswamy, C.N.; Sargent, D.J.; Velasco, R.; Maffei, M.E.; Malnoy, M. Looking forward to genetically edited fruit crops. Trends Biotechnol. 2015, 33, 62–64. [Google Scholar] [CrossRef]
- Gilbert, N.; Gewin, V.; Tollefson, J.; Sachs, J.; Potrykus, I.J.N. How to feed a hungry world. Nature 2010, 466, 531–532. [Google Scholar]
- Coleman-Derr, D.; Tringe, S.G. Building the crops of tomorrow: Advantages of symbiont-based approaches to improving abiotic stress tolerance. Front. Microbiol. 2014, 5, 283. [Google Scholar] [CrossRef]
- Christou, P. Plant genetic engineering and agricultural biotechnology 1983–2013. Trends Biotechnol. 2013, 31, 125–127. [Google Scholar] [CrossRef]
- Dong, O.X.; Ronald, P.C. Genetic Engineering for Disease Resistance in Plants: Recent Progress and Future Perspectives. Plant Physiol. 2019, 180, 26–38. [Google Scholar] [CrossRef] [Green Version]
- Giller, K.E. Genetically Engineered Crops: Experiences and Prospects; The National Academies Press: Washington, DC, USA, 2016. [Google Scholar]
- Ishii, T.; Araki, M. A future scenario of the global regulatory landscape regarding genome-edited crops. GM Crops Food 2017, 8, 44–56. [Google Scholar] [CrossRef]
- Waltz, E. With a free pass, CRISPR-edited plants reach market in record time. Nat. Biotechnol. 2018, 36, 6–7. [Google Scholar] [CrossRef]
- Wolt, J.D.; Wang, K.; Yang, B. The regulatory status of genome-edited crops. Plant Biotechnol. J. 2016, 14, 510–518. [Google Scholar] [CrossRef]
- Fritsche, S.; Poovaiah, C.; MacRae, E.; Thorlby, G. A New Zealand perspective on the application and regulation of gene editing. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef]
- Whelan, A.I.; Lema, M.A. Regulatory framework for gene editing and other new breeding techniques (NBTs) in Argentina. GM Crops Food 2015, 6, 253–265. [Google Scholar] [CrossRef] [Green Version]
- Davison, J.; Ammann, K. New GMO regulations for old: Determining a new future for EU crop biotechnology. GM Crops Food 2017, 8, 13–34. [Google Scholar] [CrossRef] [Green Version]
- Callaway, E. CRISPR plants now subject to tough GM laws in European Union. Nature 2018, 560, 16–17. [Google Scholar] [CrossRef]
| Genes | Sources | Type | Species | References |
|---|---|---|---|---|
| Attacin A (attA) | Trichoplusia ni | Antimicrobial peptide | C. sinensis cv. Hamlin/Natal/Pera/Valencia | [58,59] |
| Cecropin B and Shiva A | Synthetic | Antimicrobial peptide | C. sinensis (L.) Osbeck | [35] |
| Stx IA | Sarcophaga peregrina | Antimicrobial peptide | C. sinensis (L.) Osbeck | [60,61] |
| D2A21 | Synthetic | Antimicrobial peptide | Carrizo citrange | [62] |
| Dermaseptin gene | Synthetic | Antimicrobial peptide | C. sinensis cv. Pineapple | [63] |
| AtNPR1 | Arabidopsis thaliana | Key positive regulator of systemic acquired resistance (SAR) | C. paradisi Macf.; C. sinensis cv. Hamlin | [55] |
| CtNH1 | Citrus maxima | Key positive regulator of SAR | C. paradisi Macf. | [69] |
| Bs2 | Capsicum annuum | Resistance gene | C. sinensis cv. Hamlin/Natal/Pera/Valencia/Anliucheng; W. Murcott tangor | [72,73] |
| Xa21 | Oryza longistaminata | Resistance gene | C. limon cv. Eureka Frost Nuclear; C. sinensis cv. Pineapple | [75,76,77,78] |
| NbFLS2 | Nicotiana benthamiana | Leucine-rich repeat (LRR) receptor–like kinase gene | Carrizo citrange | [81] |
| CsMAPK1 | Citrus sinensis | Mitogen-activated protein kinase gene | Troyer citrange | [82] |
| hrpN | Erwinia amylovora | Hairpin gene | C. sinensis cv. Hamlin/Valencia | [83] |
| MdSPDS1 | Malus domestica | Spermidine synthase gene | C. sinensis cv. Anliucheng | [84] |
| terf1 | Solanum lycopersicum | Transcription factor | C. sinensis (L.) Osbeck | [85] |
| pthA-nls | Xanthomonas axonopdis pv.citri | Pathogenesis gene | C. sinensis (L.) Osbeck | [86] |
| Modified theonin | Synthetic | Cysteine-rich peptide | Carrizo citrange | [87] |
| Genes | Sources | Type | Fungi and Virus Diseases | Species | References |
|---|---|---|---|---|---|
| CTV-CP | Citrus Tristeza virus | CTV coat protein gene | Tristeza virus | C. sinensis cv. Valencia/Hamlin | [105] |
| CTV-392/393 | Citrus Tristeza virus | CTV-derived gene | C. sinensis cv. Itaborai | [106] | |
| 3DF1scFv | Hybridoma 3DF1 cell | Monoclonal antibody | C. aurantifolia (Christm.) Swing | [107] | |
| p20/23/25 | Citrus Tristeza virus | Silencing suppressor protein | C. aurantifolia (Christm.) Swing.; C. paradisi Macf. C. sinensis (L.) Osbeck | [38,39,102,103,104,109,110,116] | |
| CPsV-CP ihpCP | Citrus Psorosis virus | Coat protein gene/siRNA | Psorosis virus | C. sinensis (L.) Osbeck; | [111,112,113] |
| chit42 | Trichoderma harzianum | Endochitinase | Mal secco and gray mold | C. limon (L.) Burm. f. | [114,115] |
| PR-5 | Solanum lycopersicum | Pathogenesis-related protein | Root rot and gummosis | C. sinensis cv. Pineapple | [116] |
| Attacin E(attE) | Antimicrobial peptide | Hyalophoracecropia | Citrus scab | C. paradisi Macf. | [117] |
| CitMTSE1 | Citrus sinensis | Limonene synthase gene | Black spot | C. sinensis (L.) Osbeck | [118] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, L.; Nasrullah; Ke, F.; Nie, Z.; Wang, P.; Xu, J. Citrus Genetic Engineering for Disease Resistance: Past, Present and Future. Int. J. Mol. Sci. 2019, 20, 5256. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20215256
Sun L, Nasrullah, Ke F, Nie Z, Wang P, Xu J. Citrus Genetic Engineering for Disease Resistance: Past, Present and Future. International Journal of Molecular Sciences. 2019; 20(21):5256. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20215256
Chicago/Turabian StyleSun, Lifang, Nasrullah, Fuzhi Ke, Zhenpeng Nie, Ping Wang, and Jianguo Xu. 2019. "Citrus Genetic Engineering for Disease Resistance: Past, Present and Future" International Journal of Molecular Sciences 20, no. 21: 5256. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20215256





