Role of High-Mobility Group Box-1 in Liver Pathogenesis
Abstract
:1. HMGB1 is a Prototypical Damage-Associated Molecular Pattern Molecule
2. HMGB1 Has Multiple Functions Depending on Its Cellular Location
2.1. Nuclear HMGB1 Function as DNA Chaperone
2.2. Cytosolic HMGB1 Regulates Autophagy
2.3. Extracellular HMGB1 Functions as an Alarmin
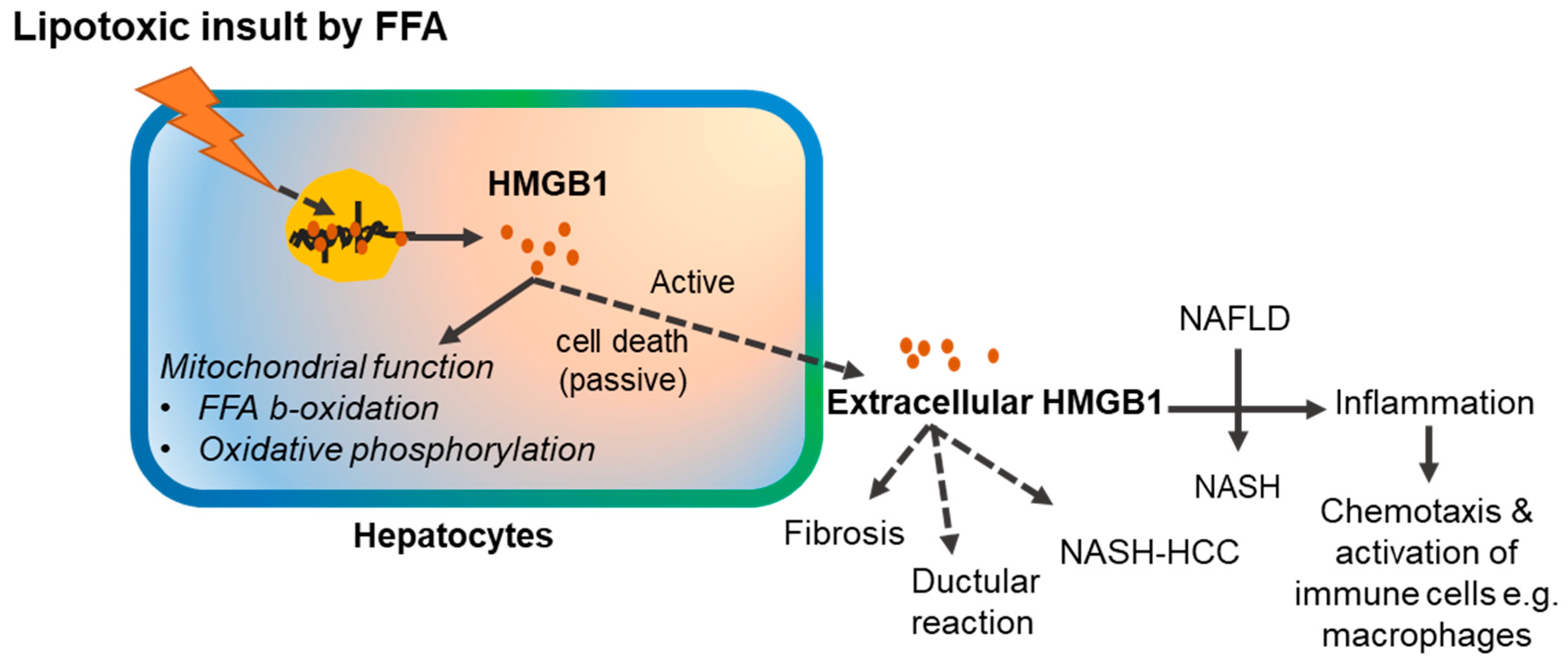
3. HMGB1 Release Depends on the Nature of Cellular Stress
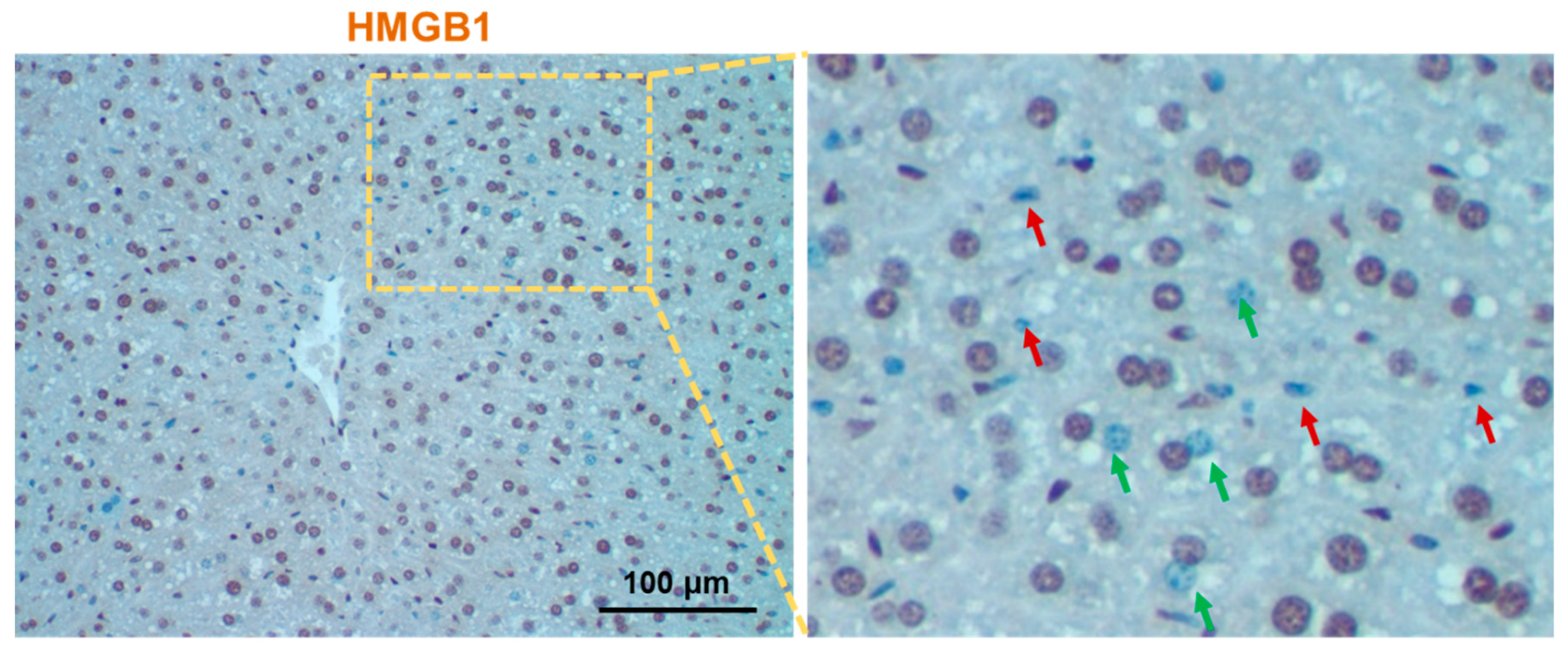
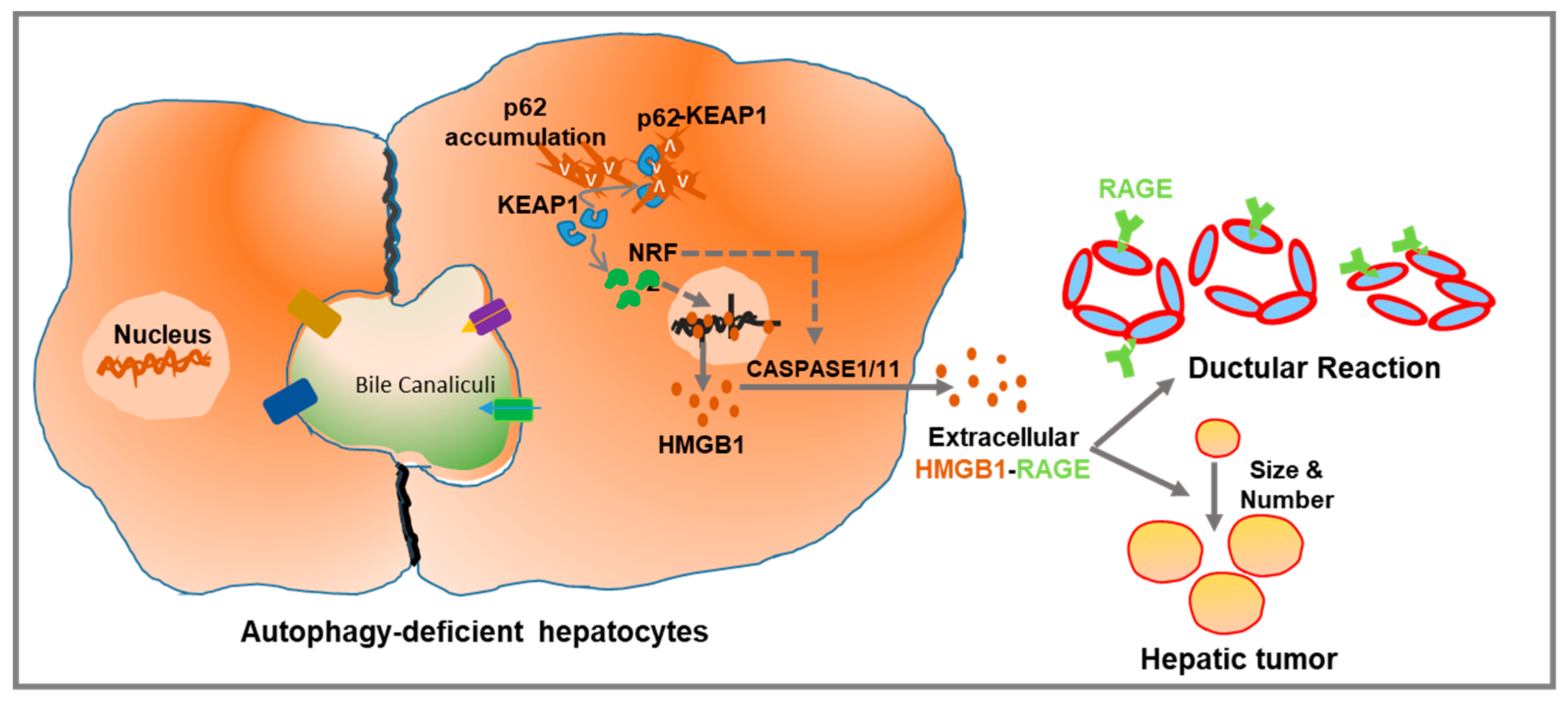
4. Hepatic Autophagy Inhibition Increases the Active Release of HMGB1
5. HMGB1 in Liver Pathogenesis
5.1. HMGB1 in Liver Inflammation
5.2. HMGB1 in Liver Fibrosis
5.3. HMGB1 in Ductular Reaction
5.4. HMGB1 in Liver Tumorigenesis
5.5. HMGB1 in Liver Regeneration
6. HMGB1 Has a Pathogenic Role in Common Liver Diseases
6.1. HMGB1 Participates in the Pathogenesis of Non-Alcoholic Fatty Liver Disease
6.2. HMGB1 Participates in the Pathogenesis of Alcoholic Liver Disease
6.3. HMGB1 in Drug-Induced Liver Injury
7. Therapeutic Potential of HMGB1
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ALD | Alcoholic liver disease |
| ANGPT1 | Angiopoietin 1 |
| APAP | Acetaminophen |
| ASH | Alcoholic steatohepatitis |
| Atg5 | Autophagy related 5 |
| Atg7 | Autophagy related 7 |
| BA | Biliary atresia |
| CCl4 | Carbon tetrachloride |
| CDE | Choline-deficient, ethionine-supplemented |
| CK19 | Cytokeratin 19 |
| CK7 | Cytokeratin 7 |
| CTGF | Connective tissue growth factor |
| DAMP | Damage-associated molecular pattern |
| DC | Ductular cell |
| DDC | 3,5-diethoxycarbonyl-1,4-dihydrocollidine |
| DEN | Diethyl nitrosamine |
| DILI | Drug-induced liver injury |
| DR | Ductular reaction |
| EpCAM | Epithelial Cell Adhesion Molecule |
| FGF10 | Fibroblast growth factor 10 |
| FGF7 | Fibroblast growth factor 7 |
| GSDMD | Gasdermin D |
| HBV | Hepatitis B virus |
| HCC | Hepatocellular carcinoma |
| HCV | Hepatitis C virus |
| HFD | High-fat diet |
| HF-HC-HSD | High-fat diet with high cholesterol and a high sugar supplement diet |
| HIF1α | Hypoxia-inducible factor 1α |
| HMGB1 | High-mobility group box 1 |
| HPC | Hepatic progenitor cell |
| HSC | Hepatic stellate cell |
| HSPB1 | Heat shock protein beta -1 |
| LGR5 | Leucine-Rich Repeat Containing G Protein-Coupled Receptor 5 |
| LPS | Lipopolysaccharide |
| MIF | Migration inhibitory factor |
| NAFLD | Non-alcoholic fatty liver disease |
| NAPQI | N-acetyl-p-benzoquinone imine |
| NASH | Non-alcoholic steatohepatitis |
| NGA2 | Neuro-Glia Antigen2 |
| NRF2 | Nuclear factor erythroid 2-related factor 2 |
| PBC | Primary biliary cirrhosis |
| PDGFC | Platelet-derived growth factor C |
| PGC-1α | Peroxisome proliferator-activated receptor γ (PPARγ) coactivator 1α |
| PSC | Primary sclerosing cholangitis |
| RAGE | Receptor for advanced glycation end products |
| RIP3 | Receptor interacting protein 3 |
| ROS | Reactive oxygen species |
| SAA | Serum Amyloid A |
| SalB | Salvianolic acid B |
| SOX9 | Sex-Determining Region Y-Box 9 |
| TLRs | Toll-like receptors |
| TRIF | TIR domain-containing adapter-inducing interferon-β |
| TROP2 | Trophoblast Cell Surface Antigen 2 |
| TWEAK | TNF-like weak inducer of apoptosis |
| VEGFD | Vascular endothelial growth factor-D |
| YAP | Yes-associated protein |
References
- Bianchi, M.; Falciola, L.; Ferrari, S.; Lilley, D. The DNA binding site of HMG1 protein is composed of two similar segments (HMG boxes), both of which have counterparts in other eukaryotic regulatory proteins. EMBO J. 1992, 11, 1055–1063. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, S.; Ronfani, L.; Calogero, S.; Bianchi, M. E The mouse gene coding for high mobility group 1 protein (HMG1). J. Boil. Chem. 1994, 269, 28803–28808. [Google Scholar]
- Huang, J.-K.; Johnson, B.H.; Gerald, R.; Wen, L. A human placental cDNA clone that encodes nonhistone chromosomal protein HMG-1. Nucleic Acids Res. 1989, 17, 1197–1214. [Google Scholar] [Green Version]
- Wang, H.; Bloom, O.; Zhang, M.; Vishnubhakat, J.M.; Ombrellino, M.; Che, J.; Frazier, A.; Yang, H.; Ivanova, S.; Borovikova, L.; et al. HMG-1 as a Late Mediator of Endotoxin Lethality in Mice. Science 1999, 285, 248–251. [Google Scholar] [CrossRef] [PubMed]
- Scaffidi, P.; Misteli, T.; Bianchi, M.E. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature 2002, 418, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Bell, C.W.; Jiang, W.; Reich, C.F., III; Pisetsky, D.S. The extracellular release of HMGB1 during apoptotic cell death. Am. J. Physiol.-Cell Physiol. 2006, 291, C1318–C1325. [Google Scholar] [CrossRef]
- Khambu, B.; Huda, N.; Chen, X.; Antoine, D.J.; Li, Y.; Dai, G.; Köhler, U.A.; Zong, W.-X.; Waguri, S.; Werner, S.; et al. HMGB1 promotes ductular reaction and tumorigenesis in autophagy-deficient livers. J. Clin. Investig. 2018, 128, 2419–2435. [Google Scholar] [CrossRef]
- Tang, Y.; Zhao, X.; Antoine, D.; Xiao, X.; Wang, H.; Andersson, U.; Billiar, T.R.; Tracey, K.J.; Lu, B. Regulation of Posttranslational Modifications of HMGB1 During Immune Responses. Antioxidants Redox Signal. 2016, 24, 620–634. [Google Scholar] [CrossRef]
- Richard, S.A.; Jiang, Y.; Xiang, L.H.; Zhou, S.; Wang, J.; Su, Z.; Xu, H. Post-translational modifications of high mobility group box 1 and cancer. Am. J. Transl. Res. 2017, 9, 5181–5196. [Google Scholar]
- Ciucci, A.; Gabriele, I.; Percario, Z.A.; Affabris, E.; Colizzi, V.; Mancino, G. HMGB1 and Cord Blood: Its Role as Immuno-Adjuvant Factor in Innate Immunity. PLoS ONE 2011, 6, e23766. [Google Scholar] [CrossRef]
- Merenmies, J.; Pihlaskari, R.; Laitinen, J.; Wartiovaara, J.; Rauvala, H. 30-kDa heparin-binding protein of brain (amphoterin) involved in neurite outgrowth. Amino acid sequence and localization in the filopodia of the advancing plasma membrane. J. Boil. Chem. 1991, 266, 16722–16729. [Google Scholar]
- Taguchi, A.; Blood, D.C.; Del Toro, G.; Canet, A.; Lee, D.C.; Qu, W.; Tanji, N.; Lu, Y.; Lalla, E.; Fu, C.; et al. Blockade of RAGE–amphoterin signalling suppresses tumour growth and metastases. Nature 2000, 405, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Falciola, L.; Spada, F.; Calogero, S.; Längst, G.; Voit, R.; Grummt, I.; Bianchi, M.E. High Mobility Group 1Protein Is Not Stably Associated with the Chromosomes of Somatic Cells. J. Cell Boil. 1997, 137, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Pil, P.M.; Chow, C.S.; Lippard, S.J. High-mobility-group 1 protein mediates DNA bending as determined by ring closures. Proc. Natl. Acad. Sci. USA 1993, 90, 9465–9469. [Google Scholar] [CrossRef] [PubMed]
- Calogero, S.; Grassi, F.; Aguzzi, A.; Voigtländer, T.; Ferrier, P.; Ferrari, S.; Bianchi, M.E. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nature Genet. 1999, 22, 276–280. [Google Scholar] [CrossRef] [PubMed]
- Huebener, P.; Gwak, G.-Y.; Pradère, J.-P.; Quinzii, C.M.; Friedman, R.; Lin, C.-S.; Trent, C.M.; Mederacke, I.; Zhao, E.; Dapito, D.H.; et al. High-mobility group box 1 is dispensable for autophagy, mitochondrial quality control, and organ function in vivo. Cell Metab. 2014, 19, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Kang, R.; Livesey, K.M.; Cheh, C.-W.; Farkas, A.; Loughran, P.; Hoppe, G.; Bianchi, M.E.; Tracey, K.J.; Zeh, H.J.; et al. Endogenous HMGB1 regulates autophagy. J. Cell Biol. 2010, 190, 881–892. [Google Scholar] [CrossRef]
- Livesey, K.M.; Kang, R.; Vernon, P.; Buchser, W.; Loughran, P.; Watkins, S.C.; Zhang, L.; Manfredi, J.J.; Zeh, H.J.; Li, L.; et al. p53/HMGB1 complexes regulate autophagy and apoptosis. Cancer Res. 2012, 72, 1996–2005. [Google Scholar] [CrossRef]
- Zhu, X.; Messer, J.S.; Wang, Y.; Lin, F.; Cham, C.M.; Chang, J.; Billiar, T.R.; Lotze, M.T.; Boone, D.L.; Chang, E.B. Cytosolic HMGB1 controls the cellular autophagy/apoptosis checkpoint during inflammation. J. Clin. Investig. 2015, 125, 1098–1110. [Google Scholar] [CrossRef] [Green Version]
- Tang, D.; Kang, R.; Livesey, K.M.; Kroemer, G.; Billiar, T.R.; Van Houten, B.; Zeh, H.J.; Lotze, M.T. High-mobility group box 1 is essential for mitochondrial quality control. Cell Metab. 2011, 13, 701–711. [Google Scholar] [CrossRef]
- Yang, H.; Wang, H.; Chavan, S.S.; Andersson, U. High Mobility Group Box Protein 1 (HMGB1): The Prototypical Endogenous Danger Molecule. Mol. Med. 2015, 21, S6–S12. [Google Scholar] [CrossRef] [PubMed]
- Arriazu, E.; Ge, X.; Leung, T.M.; Magdaleno, F.; Lopategi, A.; Lu, Y.; Kitamura, N.; Urtasun, R.; Theise, N.; Antoine, D.J.; et al. Signalling via the osteopontin and high mobility group box-1 axis drives the fibrogenic response to liver injury. Gut 2017, 66, 1123–1137. [Google Scholar] [CrossRef] [PubMed]
- Huttunen, H.J. Receptor for Advanced Glycation End Products (RAGE)-mediated Neurite Outgrowth and Activation of NF-kappa B Require the Cytoplasmic Domain of the Receptor but Different Downstream Signaling Pathways. J. Boil. Chem. 1999, 274, 19919–19924. [Google Scholar] [CrossRef] [PubMed]
- Ge, X.; Arriazu, E.; Magdaleno, F.; Antoine, D.J.; Cruz, R.D.; Theise, N.; Nieto, N. High Mobility Group Box-1 Drives Fibrosis Progression Signaling via the Receptor for Advanced Glycation End Products in Mice. Hepatology 2018, 68, 2380–2404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsung, A.; Sahai, R.; Tanaka, H.; Nakao, A.; Fink, M.P.; Lotze, M.T.; Yang, H.; Li, J.; Tracey, K.J.; Geller, D.A.; et al. The nuclear factor HMGB1 mediates hepatic injury after murine liver ischemia-reperfusion. J. Exp. Med. 2005, 201, 1135–1143. [Google Scholar] [CrossRef]
- Abeyama, K.; Stern, D.M.; Ito, Y.; Kawahara, K.-I.; Yoshimoto, Y.; Tanaka, M.; Uchimura, T.; Ida, N.; Yamazaki, Y.; Yamada, S.; et al. The N-terminal domain of thrombomodulin sequesters high-mobility group-B1 protein, a novel antiinflammatory mechanism. J. Clin. Investig. 2005, 115, 1267–1274. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Wang, H.; Levine, Y.A.; Gunasekaran, M.K.; Wang, Y.; Addorisio, M.; Zhu, S.; Li, W.; Li, J.; De Kleijn, D.P.; et al. Identification of CD163 as an antiinflammatory receptor for HMGB1-haptoglobin complexes. JCI Insight 2016, 1. [Google Scholar] [CrossRef] [Green Version]
- Janko, C.; Filipović, M.; Munoz, L.E.; Schorn, C.; Schett, G.; Ivanović-Burmazović, I.; Herrmann, M. Redox modulation of HMGB1-related signaling. Antioxid. Redox Signal. 2014, 20, 1075–1085. [Google Scholar] [CrossRef]
- Gorr, S.-U.; Darling, D.S. An N-terminal hydrophobic peak is the sorting signal of regulated secretory proteins. FEBS Lett. 1995, 361, 8–12. [Google Scholar] [CrossRef] [Green Version]
- Sims, G.P.; Rowe, D.C.; Rietdijk, S.T.; Herbst, R.; Coyle, A.J. HMGB1 and RAGE in Inflammation and Cancer. Annu. Rev. Immunol. 2010, 28, 367–388. [Google Scholar] [CrossRef]
- Hwang, J.S.; Choi, H.S.; Ham, S.A.; Yoo, T.; Lee, W.J.; Paek, K.S.; Seo, H.G. Deacetylation-mediated interaction of SIRT1-HMGB1 improves survival in a mouse model of endotoxemia. Sci. Rep. 2015, 5, 15971. [Google Scholar] [CrossRef] [PubMed]
- Evankovich, J.; Cho, S.W.; Zhang, R.; Cardinal, J.; Dhupar, R.; Zhang, L.; Klune, J.R.; Zlotnicki, J.; Billiar, T.; Tsung, A. High Mobility Group Box 1 Release from Hepatocytes during Ischemia and Reperfusion Injury Is Mediated by Decreased Histone Deacetylase Activity*. J. Boil. Chem. 2010, 285, 39888–39897. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Dupont, N.; Castillo, E.F.; Deretic, V. Secretory versus degradative autophagy: Unconventional secretion of inflammatory mediators. J. Innate Immun. 2013, 5, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Dupont, N.; Jiang, S.; Pilli, M.; Ornatowski, W.; Bhattacharya, D.; Deretic, V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1beta. EMBO J. 2011, 30, 4701–4711. [Google Scholar] [CrossRef]
- Lee, J.P.W.; Foote, A.; Fan, H.; De Castro, C.P.; Lang, T.; Jones, S.A.; Gavrilescu, N.; Mills, K.H.G.; Leech, M.; Morand, E.F.; et al. Loss of autophagy enhances MIF/macrophage migration inhibitory factor release by macrophages. Autophagy 2016, 12, 907–916. [Google Scholar] [CrossRef] [Green Version]
- Kayagaki, N.; Stowe, I.B.; Lee, B.L.; O’Rourke, K.; Anderson, K.; Warming, S.; Cuellar, T.; Haley, B.; Roose-Girma, M.; Phung, Q.T.; et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 2015, 526, 666–671. [Google Scholar] [CrossRef]
- Shi, J.; Zhao, Y.; Wang, K.; Shi, X.; Wang, Y.; Huang, H.; Zhuang, Y.; Cai, T.; Wang, F.; Shao, F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015, 526, 660–665. [Google Scholar] [CrossRef]
- Aglietti, R.A.; Estevez, A.; Gupta, A.; Ramirez, M.G.; Liu, P.S.; Kayagaki, N.; Ciferri, C.; Dixit, V.M.; Dueber, E.C. GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc. Natl. Acad. Sci. USA 2016, 113, 7858–7863. [Google Scholar] [CrossRef] [Green Version]
- Ding, J.; Wang, K.; Liu, W.; She, Y.; Sun, Q.; Shi, J.; Sun, H.; Wang, D.-C.; Shao, F. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 2016, 535, 111–116. [Google Scholar] [CrossRef]
- Yang, H.; Ochani, M.; Li, J.; Qiang, X.; Tanovic, M.; Harris, H.E.; Susarla, S.M.; Ulloa, L.; Wang, H.; DiRaimo, R.; et al. Reversing established sepsis with antagonists of endogenous high-mobility group box 1. Proc. Natl. Acad. Sci. USA 2004, 101, 296–301. [Google Scholar] [CrossRef]
- Kao, Y.-H.; Jawan, B.; Goto, S.; Hung, C.-T.; Lin, Y.-C.; Nakano, T.; Hsu, L.-W.; Lai, C.-Y.; Tai, M.-H.; Chen, C.-L. High-Mobility Group Box 1 Protein Activates Hepatic Stellate Cells in Vitro. Transplant. Proc. 2008, 40, 2704–2705. [Google Scholar] [CrossRef] [PubMed]
- Albayrak, A.; Uyanik, M.H.; Cerrah, S.; Altas, S.; Dursun, H.; Demir, M.; Uslu, H. Is HMGB1 a New Indirect Marker for Revealing Fibrosis in Chronic Hepatitis and a New Therapeutic Target in Treatment? Viral Immunol. 2010, 23, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, C.; Huebener, P.; Pradere, J.-P.; Antoine, D.J.; Friedman, R.A.; Schwabe, R.F. HMGB1 links chronic liver injury to progenitor responses and hepatocarcinogenesis. J. Clin. Investig. 2018, 128, 2436–2451. [Google Scholar] [CrossRef] [PubMed]
- Popper, H.; Kent, G.; Stein, R. Ductular cell reaction in the liver in hepatic injury. J. Mt. Sinai Hosp. N.Y. 1957, 24, 551–556. [Google Scholar] [PubMed]
- Lowes, K.N.; Brennan, B.A.; Yeoh, G.C.; Olynyk, J.K. Oval Cell Numbers in Human Chronic Liver Diseases Are Directly Related to Disease Severity. Am. J. Pathol. 1999, 154, 537–541. [Google Scholar] [CrossRef] [Green Version]
- Roskams, T.A.; Theise, N.D.; Balabaud, C.; Bhagat, G.; Bhathal, P.S.; Bioulac-Sage, P.; Brunt, E.M.; Crawford, J.M.; Crosby, H.A.; Desmet, V.; et al. Nomenclature of the finer branches of the biliary tree: Canals, ductules, and ductular reactions in human livers. Hepatology 2004, 39, 1739–1745. [Google Scholar] [CrossRef]
- Roskams, T.; Yang, S.Q.; Koteish, A.; Durnez, A.; Devos, R.; Huang, X.; Achten, R.; Verslype, C.; Diehl, A.M. Oxidative Stress and Oval Cell Accumulation in Mice and Humans with Alcoholic and Nonalcoholic Fatty Liver Disease. Am. J. Pathol. 2003, 163, 1301–1311. [Google Scholar] [CrossRef] [Green Version]
- Pusterla, T.; Németh, J.; Stein, I.; Wiechert, L.; Knigin, D.; Marhenke, S.; Longerich, T.; Kumar, V.; Arnold, B.; Vogel, A.; et al. Receptor for advanced glycation endproducts (RAGE) is a key regulator of oval cell activation and inflammation-associated liver carcinogenesis in mice. Hepatology 2013, 58, 363–373. [Google Scholar] [CrossRef]
- Utley, S.; James, D.; Mavila, N.; Nguyen, M.V.; Vendryes, C.; Salisbury, S.M.; Phan, J.; Wang, K.S. Fibroblast growth factor signaling regulates the expansion of A6-expressing hepatocytes in association with AKT-dependent beta-catenin activation. J. Hepatology 2014, 60, 1002–1009. [Google Scholar] [CrossRef]
- Jakubowski, A.; Ambrose, C.; Parr, M.; Lincecum, J.M.; Wang, M.Z.; Zheng, T.S.; Browning, B.; Michaelson, J.S.; Baestcher, M.; Wang, B.; et al. TWEAK induces liver progenitor cell proliferation. J. Clin. Investig. 2005, 115, 2330–2340. [Google Scholar] [CrossRef] [Green Version]
- Takase, H.M.; Itoh, T.; Ino, S.; Wang, T.; Koji, T.; Akira, S.; Takikawa, Y.; Miyajima, A. FGF7 is a functional niche signal required for stimulation of adult liver progenitor cells that support liver regeneration. Genes Dev. 2013, 27, 169–181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pi, L.; Robinson, P.M.; Jorgensen, M.; Oh, S.H.; Brown, A.R.; Weinreb, P.H.; Trinh, T.L.; Yianni, P.; Liu, C.; Leask, A.; et al. Connective tissue growth factor and integrin alphavbeta6: A new pair of regulators critical for ductular reaction and biliary fibrosis in mice. Hepatology 2015, 61, 678–691. [Google Scholar] [CrossRef] [PubMed]
- Michelotti, G.A.; Tucker, A.; Swiderska-Syn, M.; Machado, M.V.; Choi, S.S.; Kruger, L.; Soderblom, E.; Thompson, J.W.; Mayer-Salman, M.; Himburg, H.A.; et al. Pleiotrophin regulates the ductular reaction by controlling the migration of cells in liver progenitor niches. Gut 2016, 65, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Cheng, B.-Q.; Jia, C.-Q.; Liu, C.-T.; Lu, X.-F.; Zhong, N.; Zhang, Z.-L.; Fan, W.; Li, Y.-Q. Serum high mobility group box chromosomal protein 1 is associated with clinicopathologic features in patients with hepatocellular carcinoma. Dig. Liver Dis. 2008, 40, 446–452. [Google Scholar] [CrossRef]
- Yan, W.; Chang, Y.; Liang, X.; Cardinal, J.S.; Huang, H.; Thorne, S.H.; Monga, S.P.S.; Geller, D.A.; Lotze, M.T.; Tsung, A. High-mobility group box 1 activates caspase-1 and promotes hepatocellular carcinoma invasiveness and metastases. Hepatology 2012, 55, 1863–1875. [Google Scholar] [CrossRef]
- Dong, Y.D.; Cui, L.; Peng, C.H.; Cheng, D.F.; Han, B.S.; Huang, F. Expression and clinical significance of HMGB1 in human liver cancer: Knockdown inhibits tumor growth and metastasis in vitro and in vivo. Oncol. Rep. 2013, 29, 87–94. [Google Scholar] [CrossRef]
- Lee, S.; Zhou, P.; Gupta, A.; Shin, S. Reactive Ductules Are Associated with Angiogenesis and Tumor Cell Proliferation in Pediatric Liver Cancer. Hepatology Commun. 2018, 2, 1199–1212. [Google Scholar] [CrossRef]
- Chen, R.; Zhu, S.; Wang, H.; Lotze, M.T.; Zeh, H.J.; Billiar, T.R.; Kang, R.; Tang, D.; Fan, X.-G.; Fan, X. High mobility group protein B1 controls liver cancer initiation through yes-associated protein -dependent aerobic glycolysis. Hepatology 2018, 67, 1823–1841. [Google Scholar] [CrossRef]
- Tohme, S.; Yazdani, H.O.; Liu, Y.; Loughran, P.; Van Der Windt, D.J.; Huang, H.; Simmons, R.L.R.L.; Shiva, S.; Tai, S.; Tsung, A. Hypoxia mediates mitochondrial biogenesis in hepatocellular carcinoma to promote tumor growth through HMGB1 and TLR9 interaction. Hepatology 2017, 66, 182–197. [Google Scholar] [CrossRef] [Green Version]
- Mollica, L.; De Marchis, F.; Spitaleri, A.; Dallacosta, C.; Pennacchini, D.; Zamai, M.; Agresti, A.; Trisciuoglio, L.; Musco, G.; Bianchi, M.E. Glycyrrhizin Binds to High-Mobility Group Box 1 Protein and Inhibits Its Cytokine Activities. Chem. Boil. 2007, 14, 431–441. [Google Scholar] [CrossRef] [Green Version]
- Veldt, B.J.; Hansen, B.E.; Ikeda, K.; Verhey, E.; Suzuki, H.; Schalm, S.W. Long-term clinical outcome and effect of glycyrrhizin in 1093 chronic hepatitis C patients with non-response or relapse to interferon. Scand. J. Gastroenterol. 2006, 41, 1087–1094. [Google Scholar] [CrossRef] [PubMed]
- Karin, M.; Clevers, H. Reparative inflammation takes charge of tissue regeneration. Nature 2016, 529, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Tirone, M.; Tran, N.L.; Ceriotti, C.; Gorzanelli, A.; Canepari, M.; Bottinelli, R.; Raucci, A.; Di Maggio, S.; Santiago, C.; Mellado, M.; et al. High mobility group box 1 orchestrates tissue regeneration via CXCR4. J. Exp. Med. 2018, 215, 303–318. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, S.H.; Hirsova, P.; Gores, G.J. Non-alcoholic steatohepatitis pathogenesis: Sublethal hepatocyte injury as a driver of liver inflammation. Gut 2018, 67, 963–972. [Google Scholar] [CrossRef] [PubMed]
- Alisi, A.; Nobili, V.; Ceccarelli, S.; Panera, N.; De Stefanis, C.; De Vito, R.; Vitali, R.; Bedogni, G.; Balsano, C.; Cucchiara, S.; et al. Plasma high mobility group box 1 protein reflects fibrosis in pediatric nonalcoholic fatty liver disease. Expert Rev. Mol. Diagn. 2014, 14, 763–771. [Google Scholar] [CrossRef]
- Chen, X.; Ling, Y.; Wei, Y.; Tang, J.; Ren, Y.; Zhang, B.; Jiang, F.; Li, H.; Wang, R.; Wen, W.; et al. Dual regulation of HMGB1 by combined JNK1/2–ATF2 axis with miR-200 family in nonalcoholic steatohepatitis in mice. FASEB J. 2018, 32, 2722–2734. [Google Scholar] [CrossRef]
- Ganz, M.; Bukong, T.N.; Csak, T.; Saha, B.; Park, J.-K.; Ambade, A.; Kodys, K.; Szabo, G. Progression of non-alcoholic steatosis to steatohepatitis and fibrosis parallels cumulative accumulation of danger signals that promote inflammation and liver tumors in a high fat-cholesterol-sugar diet model in mice. J. Transl. Med. 2015, 13, 193. [Google Scholar] [CrossRef]
- Chandrashekaran, V.; Seth, R.K.; Dattaroy, D.; Alhasson, F.; Ziolenka, J.; Carson, J.; Berger, F.G.; Kalyanaraman, B.; Diehl, A.M.; Chatterjee, S. HMGB1-RAGE pathway drives peroxynitrite signaling-induced IBD-like inflammation in murine nonalcoholic fatty liver disease. Redox Boil. 2017, 13, 8–19. [Google Scholar] [CrossRef]
- Song, F.; Del Pozo, C.H.; Rosario, R.; Zou, Y.S.; Ananthakrishnan, R.; Xu, X.; Patel, P.R.; Benoit, V.M.; Yan, S.F.; Li, H.; et al. RAGE Regulates the Metabolic and Inflammatory Response to High-Fat Feeding in Mice. Diabetes 2014, 63, 1948–1965. [Google Scholar] [CrossRef] [Green Version]
- Zeng, W.; Shan, W.; Gao, L.; Gao, D.; Hu, Y.; Wang, G.; Zhang, N.; Li, Z.; Tian, X.; Xu, W.; et al. Inhibition of HMGB1 release via salvianolic acid B-mediated SIRT1 up-regulation protects rats against non-alcoholic fatty liver disease. Sci. Rep. 2015, 5, 16013. [Google Scholar] [CrossRef] [Green Version]
- Lin, M.J.; Deng, M.J.; Billiar, T.R.; Scott, M.J. Hepatocyte-specific high-mobility group box 1 (HC-HMGB1) protects against liver fat accumulation and cellular stress during high fat diet feeding. FASEB J. 2018, 32, 150–157. [Google Scholar]
- Ge, X.; Antoine, D.J.; Lu, Y.; Arriazu, E.; Leung, T.-M.; Klepper, A.L.; Branch, A.D.; Fiel, M.I.; Nieto, N. High mobility group box-1 (HMGB1) participates in the pathogenesis of alcoholic liver disease (ALD). J. Boil. Chem. 2014, 289, 22672–22691. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.M. Etiologies of acute liver failure. Semin. Liver Dis. 2008, 28, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Jollow, D.J.; Mitchell, J.R.; Potter, W.Z.; Davis, D.C.; Gillette, J.R.; Brodie, B.B. Acetaminophen-induced hepatic necrosis. II. Role of covalent binding in vivo. J. Pharmacol. Exp. Ther. 1973, 187, 195–202. [Google Scholar] [PubMed]
- Huebener, P.; Pradere, J.P.; Hernandez, C.; Gwak, G.Y.; Caviglia, J.M.; Mu, X.; Loike, J.D.; Jenkins, R.E.; Antoine, D.J.; Schwabe, R.F. The HMGB1/RAGE axis triggers neutrophil-mediated injury amplification following necrosis. J. Clin. Investig. 2015, 125, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Zamora, R.; Barclay, D.; Yin, J.; Alonso, E.M.; Leonis, M.A.; Mi, Q.; Billiar, T.R.; Simmons, R.L.; Squires, R.H.; Vodovotz, Y. HMGB1 is a Central Driver of Dynamic Pro-Inflammatory Networks in Pediatric Acute Liver Failure induced by Acetaminophen. Sci. Rep. 2019, 9, 5971. [Google Scholar] [CrossRef]
- Antoine, D.J.; Dear, J.W.; Lewis, P.S.; Platt, V.; Coyle, J.; Masson, M.; Thanacoody, R.H.; Gray, A.J.; Webb, D.J.; Moggs, J.G.; et al. Mechanistic biomarkers provide early and sensitive detection of acetaminophen-induced acute liver injury at first presentation to hospital. Hepatology 2013, 58, 777–787. [Google Scholar] [CrossRef]
- Yang, R.; Zou, X.; Tenhunen, J.; Zhu, S.; Kajander, H.; Koskinen, M.-L.; Tonnessen, T.I. HMGB1 neutralization is associated with bacterial translocation during acetaminophen hepatotoxicity. BMC Gastroenterol. 2014, 14, 66. [Google Scholar] [CrossRef]
- Wang, X.; Sun, R.; Wei, H.; Tian, Z. High-mobility group box 1 (HMGB1)-Toll-like receptor (TLR)4-interleukin (IL)-23-IL-17A axis in drug-induced damage-associated lethal hepatitis: Interaction of gammadelta T cells with macrophages. Hepatology 2013, 57, 373–384. [Google Scholar] [CrossRef]
- Kim, S.Y.; Son, M.; Lee, S.E.; Park, I.H.; Kwak, M.S.; Han, M.; Lee, H.S.; Kim, E.S.; Kim, J.-Y.; Lee, J.E.; et al. High-Mobility Group Box 1-Induced Complement Activation Causes Sterile Inflammation. Front. Immunol. 2018, 9, 705. [Google Scholar] [CrossRef] [Green Version]
- Minsart, C.; Liefferinckx, C.; Lemmers, A.; Quertinmont, E.; Dressen, C.; Goriely, S.; Moreau, R.; Gustot, T. Mo1430—Hepatocyte Release of HMGB1 During Acetaminophen-Induced Liver Injury and Deleterious Feed-Forward Process: Implication of Necroptosis Through TRIF/RIPK3 Axis. Gastroenterology 2018, 154, S-1204. [Google Scholar] [CrossRef]
- Yan, T.; Wang, H.; Zhao, M.; Yagai, T.; Chai, Y.; Krausz, K.W.; Xie, C.; Cheng, X.; Zhang, J.; Che, Y.; et al. Glycyrrhizin Protects against Acetaminophen-Induced Acute Liver Injury via Alleviating Tumor Necrosis Factor Alpha-Mediated Apoptosis. Drug Metab. Dispos. 2016, 44, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Zhang, S.; Cotoia, A.; Oksala, N.; Zhu, S.; Tenhunen, J. High mobility group B1 impairs hepatocyte regeneration in acetaminophen hepatotoxicity. BMC Gastroenterol. 2012, 12, 45. [Google Scholar] [CrossRef] [PubMed]
- VanPatten, S.; Al-Abed, Y. High Mobility Group Box-1 (HMGb1): Current Wisdom and Advancement as a Potential Drug Target. J. Med. Chem. 2018, 61, 5093–5107. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khambu, B.; Yan, S.; Huda, N.; Yin, X.-M. Role of High-Mobility Group Box-1 in Liver Pathogenesis. Int. J. Mol. Sci. 2019, 20, 5314. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20215314
Khambu B, Yan S, Huda N, Yin X-M. Role of High-Mobility Group Box-1 in Liver Pathogenesis. International Journal of Molecular Sciences. 2019; 20(21):5314. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20215314
Chicago/Turabian StyleKhambu, Bilon, Shengmin Yan, Nazmul Huda, and Xiao-Ming Yin. 2019. "Role of High-Mobility Group Box-1 in Liver Pathogenesis" International Journal of Molecular Sciences 20, no. 21: 5314. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20215314




