Identification and Characterization of a Fatty Acid- and Retinoid-Binding Protein Gene (Ar-far-1) from the Chrysanthemum Foliar Nematode, Aphelenchoides ritzemabosi
Abstract
:1. Introduction
2. Results
2.1. Cloning and Analysis of Full-Length Ar-far-1 from CFN
2.2. Southern Blot Analysis
2.3. Ligand Binding
2.4. Localization and Expression of Ar-far-1 mRNA
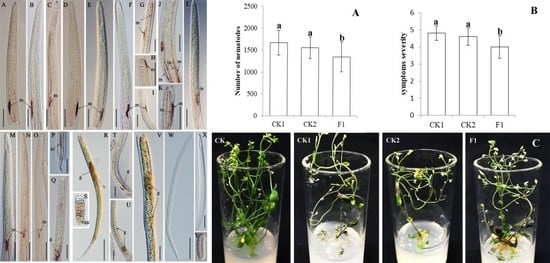
2.5. Ar-far-1 Silencing Efficiency and Influence on the Reproduction and Pathogenicity of CFN
3. Discussion
4. Materials and Methods
4.1. Nematodes
4.2. RNA Extraction and Cloning of Ar-far-1 from CFN
4.3. Sequence Analysis, Alignment and Phylogenetics
4.4. Southern Blot Hybridization
4.5. Expression and Purification of Recombinant Ar-FAR-1 and Fluorescence-Based Ligand-Binding Assays
4.6. In Situ Hybridization
4.7. Expression of Ar-far-1 mRNA at Different Developmental Stages of CFN
4.8. Reproduction and Pathogenicity of CFN after RNAi Treatment
4.9. Data Analysis
Data Availability Statement
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siddiqi, M. Aphelenchoides ritzemabosi. In CIH Descriptions of Plant Parasitic Nematodes; Set 3, No.32; CAB International: Wallingford, UK, 1974; p. 4. [Google Scholar]
- Wang, D.W.; Peng, X.F.; Xie, H.; Xu, C.L.; Cheng, D.Q.; Li, J.Y.; Wu, W.J.; Wang, K. Arabidopsis thaliana as a suitable model host for research on interactions between plant and foliar nematodes, parasites of plant shoot. Sci. Rep. 2016, 6, 38286. [Google Scholar] [CrossRef] [Green Version]
- Xie, H. The epidemic and detection methods of Aphelenchoides ritzemabosi. Plant Quar. 2007, 21, 190–192. [Google Scholar]
- Crozzoli, R.; Hurtado, T.; Perichi, G.; Arcia, A. Characterization of a Venezuelan population of Aphelenchoides ritzemabosi on chrysanthemum. Nematol. Mediterr. 2008, 36, 79–83. [Google Scholar]
- Van Megen, H.; van den Elsen, S.; Holterman, M.; Karssen, G.; Mooyman, P.; Bongers, T.; Holovachov, O.; Bakker, J.; Helder, J. A phylogenetic tree of nematodes based on about 1200 full-length small subunit ribosomal DNA sequences. Nematology 2009, 11, 927–950. [Google Scholar] [CrossRef]
- Rybarczyk-Mydlowska, K.; Mooyman, P.; van Megen, N.; van den Elsen, S.; Vervoort, M.; Veenhuizen, P.; van Doorn, J.; Dees, R.; Karssen, G.; Bakker, J.; et al. Small Subunit Ribosomal DNA-Based Phylogenetic Analysis of Foliar Nematodes (Aphelenchoides spp.) and Their Quantitative Detection in Complex DNA Backgrounds. Phytopathology 2012, 102, 1153–1160. [Google Scholar] [CrossRef]
- Xiang, Y.; Wang, D.W.; Li, J.Y.; Xie, H.; Xu, C.L.; Li, Y. Transcriptome Analysis of the Chrysanthemum Foliar Nematode, Aphelenchoides ritzemabosi (Aphelenchida: Aphelenchoididae). PLoS ONE 2016, 11, e0166877. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, A.; Kennedy, M.W.; Bradley, J.E. The FAR proteins of parasitic nematodes: Their possible involvement in the pathogenesis of infection and the use of Caenorhabditis elegans as a model system to evaluate their function. Med. Microbiol. Immun. 2003, 192, 47–52. [Google Scholar] [CrossRef]
- Garofalo, A.; Rowlinson, M.C.; Amambua, N.A.; Hughes, J.M.; Kelly, S.M.; Price, N.C.; Cooper, A.; Watson, D.G.; Kennedy, M.W.; Bradley, J.E. The FAR protein family of the nematode Caenorhabditis elegans. Differential lipid binding properties, structural characteristics, and developmental regulation. J. Biol. Chem. 2003, 278, 8065–8074. [Google Scholar] [CrossRef]
- Prior, A.; Jones, J.T.; Blok, V.C.; Beauchamp, J.; McDermott, L.; Cooper, A.; Kennedy, M.W. A surface-associated retinol- and fatty acid-binding protein (Gp-FAR-1) from the potato cyst nematode Globodera pallida: Lipid binding activities, structural analysis and expression pattern. Biochem. J. 2001, 356, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Iberkleid, I.; Sela, N.; Miyara, S.B. Meloidogyne javanica fatty acid- and retinol-binding protein (Mj-FAR-1) regulates expression of lipid-, cell wall-, stress- and phenylpropanoid-related genes during nematode infection of tomato. BMC Genom. 2015, 16, 272. [Google Scholar] [CrossRef]
- Qiao, F.; Luo, L.; Peng, H.; Luo, S.; Huang, W.; Cui, J. Characterization of Three Novel Fatty Acid-and Retinoid-Binding Protein Genes (Ha-far-1, Ha-far-2 and Hf-far-1) from the Cereal Cyst Nematodes Heterodera avenae and H. filipjevi. PLoS ONE 2016, 11, e0160003. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Xie, H.; Cheng, X.; Wang, D.W.; Li, Y.; Xu, C.L.; Huang, X. Molecular identification and functional characterization of the fatty acid-and retinoid-binding protein gene Rs-far-1 in the burrowing nematode Radopholus similis (Tylenchida: Pratylenchidae). PLoS ONE 2015, 10, e0118414. [Google Scholar] [CrossRef]
- Basavaraju, S.V.; Zhan, B.; Kennedy, M.W.; Liu, Y.; Hawdon, J.; Hotez, P.J. Ac-FAR-1, a 20 kDa fatty acid- and retinol-binding protein secreted by adult Ancylostoma caninum hookworms: Gene transcription pattern, ligand binding properties and structural characterisation. Mol. Biochem. Parasitol. 2003, 126, 63–71. [Google Scholar] [CrossRef]
- Cheng, X.; Xiang, Y.; Xie, H.; Xu, C.L.; Xie, T.F.; Zhang, C.; Li, Y. Molecular Characterization and Functions of Fatty Acid and Retinoid Binding Protein Gene (Ab-far-1) in Aphelenchoides besseyi. PLoS ONE 2013, 8, e66011. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, M.W.; Garside, L.H.; Goodrick, L.E.; McDermott, L.; Brass, A.; Price, N.C.; Kelly, S.M.; Cooper, A.; Bradley, J.E. The Ov20 protein of the parasitic nematode Onchocerca volvulus. A structurally novel class of small helix-rich retinol-binding proteins. J. Biol. Chem. 1997, 272, 29442–29448. [Google Scholar] [CrossRef] [PubMed]
- McDermott, L.; Cooper, A.; Kennedy, M.W. Novel classes of fatty acid and retinol binding protein from nematodes. Mol. Cell Biochem. 1999, 192, 69–75. [Google Scholar] [CrossRef]
- Garofalo, A.; Klager, S.L.; Rowlinson, M.C.; Nirmalan, N.; Klion, A.; Allen, J.E.; Kennedy, M.W.; Bradley, J.E. The FAR proteins of filarial nematodes: Secretion, glycosylation and lipid binding characteristics. Mol. Biochem. Parasit. 2002, 122, 161–170. [Google Scholar] [CrossRef]
- Solovyova, A.S.; Meenan, N.; McDermott, L.; Garofalo, A.; Bradley, J.E.; Kennedy, M.W.; Byron, O. The polyprotein and FAR lipid binding proteins of nematodes: Shape and monomer/dimer states in ligand-free and bound forms. Eur. Biophys J. Biophy. 2003, 32, 465–476. [Google Scholar] [CrossRef]
- Stein, L.D.; Bao, Z.R.; Blasiar, D.; Blumenthal, T.; Brent, M.R.; Chen, N.S.; Chinwalla, A.; Clarke, L.; Clee, C.; Coghlan, A.; et al. The genome sequence of Caenorhabditis briggsae: A platform for comparative genomics. PLoS Biol. 2003, 1, e45. [Google Scholar] [CrossRef]
- Fairfax, K.C.; Vermeire, J.J.; Harrison, L.M.; Bungiro, R.D.; Grant, W.; Husain, S.Z.; Cappello, M. Characterisation of a fatty acid and retinol binding protein orthologue from the hookworm Ancylostoma ceylanicum. Int. J. Parasitol. 2009, 39, 1561–1571. [Google Scholar] [CrossRef] [PubMed]
- Iberkleid, I.; Vieira, P.; Engler, J.D.; Firester, K.; Spiegel, Y.; Horowitz, S.B. Fatty Acid-and Retinol-Binding Protein, Mj-FAR-1 Induces Tomato Host Susceptibility to Root-Knot Nematodes. PLoS ONE 2013, 8, e64586. [Google Scholar] [CrossRef] [PubMed]
- Duarte, A.; Curtis, R.; Maleita, C.; Tiago, I.; Abrantes, I. Characterization of the venom allergen-like protein (vap-1) and the fatty acid and retinol binding protein (far-1) genes in Meloidogyne hispanica. Eur. J. Plant Pathol. 2014, 139, 825–836. [Google Scholar] [CrossRef]
- Rey-Burusco, M.F.; Ibanez-Shimabukuro, M.; Gabrielsen, M.; Franchini, G.R.; Roe, A.J.; Griffiths, K.; Zhan, B.; Cooper, A.; Kennedy, M.W.; Corsico, B.; et al. Diversity in the structures and ligand-binding sites of nematode fatty acid and retinol-binding proteins revealed by Na-FAR-1 from Necator americanus. Biochem. J. 2015, 471, 403–414. [Google Scholar] [CrossRef]
- Le, X.H.; Wang, X.; Guan, T.L.; Ju, Y.L.; Li, H.M. Isolation and characterization of a fatty acid- and retinoid-binding protein from the cereal cyst nematode Heterodera avenae. Exp. Parasitol. 2016, 167, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Phani, V.; Shivakumara, T.N.; Davies, K.G.; Rao, U. Meloidogyne incognita Fatty Acid- and Retinol- Binding Protein (Mi-FAR-1) Affects Nematode Infection of Plant Roots and the Attachment of Pasteuria penetrans Endospores. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Wang, D.W.; Xu, C.L.; Ding, S.W.; Huang, X.; Cheng, X.; Zhang, C.; Chen, C.; Xie, H. Identification and function of FAR protein family genes from a transcriptome analysis of Aphelenchoides besseyi. Bioinformatics 2018, 34, 2936–2943. [Google Scholar] [CrossRef] [PubMed]
- Cozzone, A.J. Protein phosphorylation in prokaryotes. Annu. Rev. Microbiol. 1988, 42, 97–125. [Google Scholar] [CrossRef] [PubMed]
- Stock, J.B.; Ninfa, A.J.; Stock, A.M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 1989, 53, 450–490. [Google Scholar] [PubMed]
- Varki, A.; Etzler, M.E.; Cummings, R.D.; Esko, J.D. Discovery and Classification of Glycan-Binding Proteins. Essent. Glycobiol. 2009, 2. [Google Scholar]
- Urwin, P.E.; Lilley, C.J.; Atkinson, H.J. Ingestion of double-stranded RNA by preparasitic juvenile cyst nematodes leads to RNA interference. Mol. Plant Microbe Interact. 2002, 15, 747–752. [Google Scholar] [CrossRef]
- Cottrell, T.R.; Doering, T.L. Silence of the strands: RNA interference in eukaryotic pathogens. Trends Microbiol. 2003, 11, 37–43. [Google Scholar] [CrossRef]
- Chen, Q.; Rehman, S.; Smant, G.; Jones, J.T. Functional analysis of pathogenicity proteins of the potato cyst nematode Globodera rostochiensis using RNAi. Mol. Plant Microbe 2005, 18, 621–625. [Google Scholar] [CrossRef] [PubMed]
- Rosso, M.N.; Dubrana, M.P.; Cimbolini, N.; Jaubert, S.; Abad, P. Application of RNA interference to root-knot nematode genes encoding esophageal gland proteins. Mol. Plant Microbe Interact. 2005, 18, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, K.; Xie, H.; Wang, Y.T.; Wang, D.W.; Xu, C.L.; Huang, X.; Wang, D.S. A Nematode Calreticulin, Rs-CRT, Is a Key Effector in Reproduction and Pathogenicity of Radopholus similis. PLoS ONE 2015, 10, e0129351. [Google Scholar] [CrossRef]
- Reise, R.W.; Huettel, R.N.; Sayre, R.M. Carrot callus tissue for culture of endoparasitic nematodes. J. Nematol. 1987, 19, 387–389. [Google Scholar]
- Petersen, T.N.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods 2011, 8, 785–786. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Wang, K.; Li, Y.; Huang, X.; Wang, D.W.; Xu, C.L.; Xie, H. The cathepsin S cysteine proteinase of the burrowing nematode Radopholus similis is essential for the reproduction and invasion. Cell Biosci. 2016, 6, 39. [Google Scholar] [CrossRef]
- Li, X.D.; Zhuo, K.; Luo, M.; Sun, L.H.; Liao, J.L. Molecular cloning and characterization of a calreticulin cDNA from the pinewood nematode Bursaphelenchus xylophilus. Exp. Parasitol. 2011, 128, 121–126. [Google Scholar] [CrossRef]
- Cogan, U.; Kopelman, M.; Mokady, S.; Shinitzky, M. Binding affinities of retinol and related compounds to retinol binding proteins. Eur. J. Biochem. 1976, 65, 71–78. [Google Scholar] [CrossRef]
- De Boer, J.M.; Yan, Y.; Smant, G.; Davis, E.L.; Baum, T.J. In-situ Hybridization to Messenger RNA in Heterodera glycines. J. Nematol. 1998, 30, 309–312. [Google Scholar] [PubMed]
- Van Rij, R.P.; Andino, R. RNAi-A guide to gene silencing. Science 2004, 303, 1978–1979. [Google Scholar] [CrossRef]
- Zhang, C.; Xie, H.; Xu, C.L.; Cheng, X.; Li, K.M.; Li, Y. Differential expression of Rs-eng-1b in two populations of Radopholus similis (Tylenchida: Pratylecnchidae) and its relationship to pathogenicity. Eur. J. Plant Pathol. 2012, 133, 899–910. [Google Scholar] [CrossRef]








| Primer Name | Sequence | Primer Use |
|---|---|---|
| FAR1-F | 5′-AACCCAAGTTTGAGCAACCTC-3′ | cDNA and gDNA amplification |
| FAR1-R | 5′-TATGAAGTTTATTTCGTGTTT-3′ | |
| FAR1-Sb-F | 5′-AGAAGTTGTGCCCGAGGAAGT-3′ | Southern blot |
| FAR1-Sb-R | 5′-ACCTTCAACAAATGCTTTCGC-3′ | |
| FAR1FBamHI | 5′-CGGGATCCATGAGTCTTCGCACATTCGTT-3′ | Prokaryotic expression |
| FAR1RXhoI | 5′-CCGCTCGAGTTAATTTTCTTGTTTGATCAA-3′ | |
| FAR1-IN-T7F | 5′-GGATCCTAATACGACTCACTATAGGGTACTCTTCCGTTGAGCATTC-3′ | In situ hybridization and dsRNA template |
| FAR1-IN-R | 5′- GCTTCTCTTCACCCTTAGGT-3′ | |
| FAR1-IN-F | 5′- TACTCTTCCGTTGAGCATTC-3′ | |
| FAR1-IN-T7R | 5′-GGATCCTAATACGACTCACTATAGGGGCTTCTCTTCACCCTTAGGT-3′ | |
| qFAR1-F | 5′-GTCTTCGCACATTCGTTTC-3′ | qRT-PCR |
| qFAR1-R | 5′-ATTTTTTCACTTCCTCGGG-3′ | |
| Y18s-F | 5′-GACTCAACACGGGAAACCTCA-3′ | qRT-PCR |
| Y18s-R | 5′-GCAGACACTCCACACAAGCAC-3′ |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, S.-W.; Wang, D.-W.; Xiang, Y.; Xu, C.-L.; Xie, H. Identification and Characterization of a Fatty Acid- and Retinoid-Binding Protein Gene (Ar-far-1) from the Chrysanthemum Foliar Nematode, Aphelenchoides ritzemabosi. Int. J. Mol. Sci. 2019, 20, 5566. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20225566
Ding S-W, Wang D-W, Xiang Y, Xu C-L, Xie H. Identification and Characterization of a Fatty Acid- and Retinoid-Binding Protein Gene (Ar-far-1) from the Chrysanthemum Foliar Nematode, Aphelenchoides ritzemabosi. International Journal of Molecular Sciences. 2019; 20(22):5566. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20225566
Chicago/Turabian StyleDing, Shan-Wen, Dong-Wei Wang, Yu Xiang, Chun-Ling Xu, and Hui Xie. 2019. "Identification and Characterization of a Fatty Acid- and Retinoid-Binding Protein Gene (Ar-far-1) from the Chrysanthemum Foliar Nematode, Aphelenchoides ritzemabosi" International Journal of Molecular Sciences 20, no. 22: 5566. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20225566