Disclosure of the Molecular Mechanism of Wheat Leaf Spot Disease Caused by Bipolaris sorokiniana through Comparative Transcriptome and Metabolomics Analysis
Abstract
:1. Introduction
2. Results
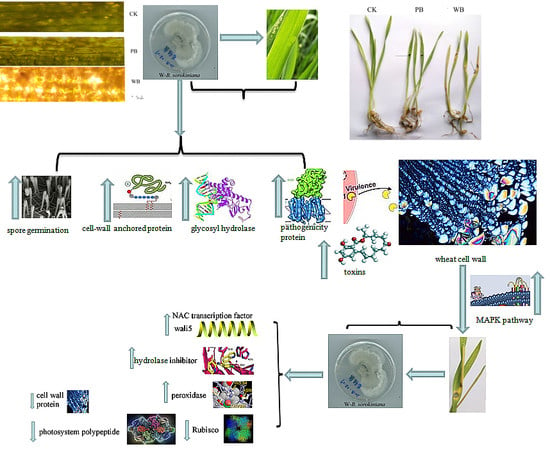
2.1. Pathogenicity of the Two B. sorokiniana Strains
2.2. The Metabolomics Analysis of the Two B. sorokiniana Strains
2.3. Annotation of the Assembled Unigenes in Transcriptomes of B. sorokiniana-Infected Wheat
2.4. The Differential Expression of Pathogenicity-Related Unigenes
2.5. Unigenes Encoding Cell Wall-Degrading Enzymes
2.6. Unigenes Related to Toxin Production
2.7. The Validation of Differential Unigenes in CK, PB and WB Groups
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Strains
4.2. Pathogenicity Testing of the Two B. sorokiniana Strains
4.3. Total RNA Extraction and Sample Preparation for RNA-Seq
4.4. Assembly, Comparative Analysis, and Functional Annotation of the Transcriptome
4.5. GO Classification and Pathway Enrichment Analyses
4.6. Comparative Expression Analysis
4.7. Validation of the Gene Expression
4.8. The Metabolomics Analysis of the Two B. sorokiniana Strains
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bailey, K.L.; Gossen, B.D.; Lafond, G.P.; Watson, P.R.; Derksen, D.A. Effect of tillage and crop rotation on root and foliar diseases of wheat and pea in Saskatchewan from 1991 to 1998: Univariate and multivariate analyses. Can. J. Plant Sci. 2001, 81, 789–803. [Google Scholar] [CrossRef]
- Adlakha, K.L.; Wilcoxson, R.D.; Raychauduri, S.P. Resistance of wheat to spot blotch caused by Bipolaris sorokiniana. Plant Dis. 1984, 68, 320–321. [Google Scholar] [CrossRef] [Green Version]
- Duveiller, E.; Altamirano, I.G. Pathogenicity of Bipolaris sorokiniana isolates from wheat roots, leaves and grains in Mexico. Plant Pathol. 2000, 49, 235–242. [Google Scholar] [CrossRef]
- Jagdish, K.; Patrick, S.; Ralph, H.; Gregor, L.; Helmut, B.; Elke, S.; Subramanism, N.; Karl-Heinz, K. Bipolaris sorokiniana, a cereal pathogen of global concern: Cytological and molecular approaches towards better control. Mol. Plant Pathol. 2002, 3, 185–195. [Google Scholar]
- Kumar, U.; Joshi, A.K.; Kumar, S.; Chard, R.; Roder, M.S. Quantitative trait loci for resistance to spot blotch caused by Bipolaris sorokiniana in wheat (T. aestivum L.) lines “Ning 8201” and “Chirya 3”. Mol. Breed. 2010, 26, 477–491. [Google Scholar] [CrossRef]
- Aggarwal, R.; Gupta, S.; Singh, V.B.; Sharma, S. Microbial detoxification of pathotoxin produced by spot blotch pathogen Bipolaris sorokiniana infecting wheat. J. Plant Biochem. Biol. 2011, 20, 66–73. [Google Scholar] [CrossRef]
- Eaton, C.J.; Cox, M.P.; Scott, B. What triggers grass endophytes to switch from mutualism to pathogenism? Plant Sci. 2011, 180, 190–195. [Google Scholar] [CrossRef]
- Schulz, B.; Boyle, C. The endophytic continuum. Mycol. Res. 2005, 109, 661–686. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.M.; Zhu, H.M.; Wen, H. A role of MAPK pathway in the pathogenic mechanism of Cryptcococcus neoformans. Chin. J. Mycol. 2012, 5, 304–308. [Google Scholar]
- Li, D.L.; Chen, Y.C.; Pan, Q.L.; Tao, M.H.; Zhang, W.M. A new eudesmane sesquiterpene from Nigrospora oryzae, an endophytic fungus of Aquilaria sinensis. Rec. Nat. Prod. 2014, 8, 330–333. [Google Scholar]
- Tao, M.H.; Chen, Y.C.; Wei, X.Y.; Tan, J.W.; Zhang, W.M. The chemical constituents of endophytic fungus Phomopsis sp. A240 isolated from Taxus chinensis var. mairei. Helv. Chim. Acta 2014, 97, 426–430. [Google Scholar] [CrossRef]
- Rodriguez, R.J.; White, J.F.; Arnold, A.E.; Redman, R.S. Fungal endophytes: Diversity and functional roles. N. Phytol. 2009, 182, 314–330. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.H.; Wang, C.; Li, S.X.; Su, Z.Z.; Zhou, H.N.; Mao, L.J.; Feng, X.X.; Liu, P.P.; Chen, X.; Snyder, J.H.; et al. Friend or foe: Differential responses of rice to invasion by mutualistic or pathogenic fungi revealed by RNAseq and metabolite profiling. Sci. Rep. 2015, 5, 13624. [Google Scholar] [CrossRef] [Green Version]
- Affokpon, A.; Coyne, D.L.; Htay, C.C.; Agbede, R.D.; Louis, L.; Coosemans, J. Biocontrol potential of native Trichoderma isolates against root-knot nematodes in West African vegetable production systems. Soil Biol. Biochem. 2011, 43, 600–608. [Google Scholar] [CrossRef]
- Dong, L.Q.; Zhang, K.Q. Micorbial control of plant-parasitic nematodes: A five-party interaction. Plant Soil 2006, 288, 31–45. [Google Scholar] [CrossRef]
- Djame, A.; Kahmann, R. Ustilago maydis: Dissecting the molecular interface between pathogen and plant. PLoS Pathol. 2012, 8, e1002955. [Google Scholar] [CrossRef] [Green Version]
- Guimil, S.; Chang, H.S.; Zhu, T.; Sesma, A.; Osbourn, A.; Roux, C. Comparative transcriptomics of rice reveals an ancient pattern of response to microbial colonization. Proc. Natl. Acad. Sci. USA 2005, 102, 8066–8070. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.H.; Su, Z.Z.; Wang, C.; Kubicek, C.P.; Feng, X.X.; Mao, L.J.; Wang, J.Y.; Chen, C.; Lin, F.C. The rice endophyte Harpophora oryzae genome reveals evolution from a pathogen to a mutualistic endophyte. Sci. Rep. 2014, 4, 5783. [Google Scholar] [CrossRef] [Green Version]
- Wikee, S.; Lorenzo, L.; Crous, P.W.; Nakashima, C.; Motohashi, K.; Chukeatirote, E.; Siti, A.A.; Eric, H.C.M.; Kevin, D.H. Phyllosticta capitalensis, a widespread endophyte of plants. Fungal Divers. 2013, 60, 91–105. [Google Scholar] [CrossRef]
- Hennings, P.; Fungi, S. Paulenses IV a cl. Puttemans collecti. Hedwigia 1908, 48, 1–20. [Google Scholar]
- Talbot, N.J.; Ebbole, D.J.; Hamer, J.E. Identification and characterization of MPG1, a gene involved in pathogenicty from the rice blast fungus Magnaporthe grisea. Plant Cell 1993, 5, 1575–1590. [Google Scholar] [PubMed] [Green Version]
- Lau, G.; Hamer, J.E. Regulatory genes controlling MPG1 expression and pathogenicity in the rice blast fungus Magnaporthe grisea. Plant Cell 1996, 8, 771–781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Z.; Zhu, L.; Song, T.; Wang, Y.; Zhang, Q.; Xia, Y.Q.; Qiu, M.; Lin, Y.; Li, H.; Kong, L.; et al. Paralogous decoy protects Phytophthora sojae apoplastic effector PsXEG1 from a host inhibitor. Science 2017, 355, 710–714. [Google Scholar] [CrossRef] [PubMed]
- Apoga, D.; Akesson, H.; Jansson, H.B.; Odham, G. Relationship between production of the phytotoxin prehelminthosporol and virulence in isolates of the plant pathogenic fungus Bipolaris sorokiniana. Plant Pathol. 2002, 108, 519–526. [Google Scholar] [CrossRef]
- Wang, M.; Sun, Z.H.; Chen, Y.C.; Liu, H.X.; Li, H.H.; Tan, G.H.; Li, S.N.; Guo, X.L.; Zhang, W.M. Cytotoxic cochlioquinone derivatives from the endophytic fungus Biopolaris sorokiniana derived from Pogostemon cablin. Fitoterapia 2016, 110, 77–82. [Google Scholar] [CrossRef]
- Xu, H.; Mendgen, K. Targeted cell wall degradation at the penetration site of cowpea rust basidiosporelings. Mol. Plant Microbe Interact. 1997, 10, 87–94. [Google Scholar] [CrossRef] [Green Version]
- Fetch, T.G.; Steffenson, B.J. Rating scales for assessing infection responses of barley infected with Cochliobolus sativus. Plant Dis. 1999, 3, 213–217. [Google Scholar] [CrossRef] [Green Version]
- Deising, H.B.; Werner, S. The role of fungal appressoria in plant infection. Microbes Infect. 2009, 2, 282–285. [Google Scholar]
- Vincent, P.; Goubet, F.; Carapito, R.; Jeltsch, J.M. Plant cell wall degradation with a powerful Fusarium graminearum Enzymatic Arsenal. J. Microbiol. Biotechnol. 2009, 19, 573–581. [Google Scholar] [CrossRef]
- Fischer, R.L.; Bennett, A.B. Role of cell wall hydrolyse in fruit ripening. Annual Reviews on. Plant Biol. 1991, 42, 675–703. [Google Scholar]
- Kikot, G.E.; Hours, R.A.; Teresa, M.A. Contribution of cell wall degrading enzymes to pathogenesis of Fusarium graminearum: A review. J. Basic Microb. 2009, 49, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Steinmetz, W.E.; Robustelli, P.; Edens, E.; Heineman, D. Structure and conformational dynamics of trichothecene mycotoxins. J. Nat. Prod. 2008, 71, 589–594. [Google Scholar] [CrossRef] [PubMed]
- Ward, T.J.; Bielawski, J.P.; Kistler, H.C.; Sullivan, E.; O’Donnell, K. Ancestral polymorphism and adaptive evolution in the trichothecene mycotoxin gene cluster of phytopathogenic Fusarium. Proc. Natl. Acad. Sci. USA 2002, 99, 9278–9283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuzick, A.; Urban, M.; Kosack, K.H. Fusarium graminearum gene deletion mutants map1 and tri5 reveal similarities and differences in the pathogenicity requirements to cause disease on Arabidopsis and wheat floral tissue. N. Phytol. 2008, 177, 990–1000. [Google Scholar] [CrossRef]
- Kojima, K.; Bahn, Y.S.; Heitman, J. Calcineurin, Mpk1 and Hog1 MAPK pathways independently control fludioxonil antifungal sensitivity in Cryptococcus neoformans. Microbiology 2006, 152, 591–604. [Google Scholar] [CrossRef] [Green Version]
- Aich, S.; Singh, R.K.; Kundu, P.; Pandey, S.P.; Datta, S. Genome-wide characterizationof cellulases from the hemi-biotrophic plant pathogen, Bipolaris sorokiniana, reveals thepresence of a highly stable GH7 endoglucanase. Biotechnol. Biofuel. 2017, 10, 135–148. [Google Scholar] [CrossRef]
- Wang, D.; Huang, X.; Zhao, L.L.; Zhao, J.J. Advances in the MAPK signaling pathway of Candida albicans. Chin. J. Mycol. 2013, 8, 252–258. [Google Scholar]
- Garg, B.; Puranik, S.; Tuteja, N.; Prasad, M. Abiotic stress-responsive expression of wali1 and wali5 genes from wheat. Plant Signal Behav. 2012, 7, 1393–1396. [Google Scholar] [CrossRef] [Green Version]
- Nakashima, K.; Tran, L.S.; Van, N.D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi-shinozaki, K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef]
- Seo, P.J.; Kim, M.J.; Park, J.Y.; Kim, S.Y.; Jeon, J.; Lee, Y.H.; Kim, J.; Park, C.M. Cold activation of a plasma membrane-tethered NAC transcription factor induces a pathogen resistance response in Arabidopsis. Plant J. 2010, 61, 661–671. [Google Scholar] [CrossRef]
- Brumbarova, T.; Matros, A.; Mock, H.P.; Bauer, P. A proteomic study showing differential regulation of stress, redox regulation and peroxidase proteins by iron supply and the transcription factor FER. Plant. J. 2008, 54, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Deng, W.Q.; Li, T.H.; Song, B.; Shen, Y.H. Illumina-based de novo transcriptome sequencing and analysis of Amanita exitialis basidiocarps. Gene 2013, 532, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Zhang, W.; Liu, T.; Huang, Z.; Zhu, M.; Chen, Y.; Li, H.H.; Li, S.N. De novo transcriptome sequencing of the deep-sea-derived fungus Dichotomomyces cejpii and analysis of gliotoxin biosynthesis genes. Int. J. Mol. Sci. 2018, 19, 1910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Yu, X.; Ma, F.; Li, J. RNA-Seq analysis of transcriptome and glucosinolate metabolism in seeds and sprouts of Broccoli (Brassica oleracea var. italica). PLoS ONE 2014, 9, e88804. [Google Scholar]
- Wu, J.K.; Zhang, W.Q.; Huang, S.B.; He, Z.Q.; Cheng, Y.B.; Wang, J. SOAP fusion: A robust and effective computational fusion discovery tool for RNA-seq reads. Bioinformatics 2013, 29, 8. [Google Scholar] [CrossRef] [Green Version]
- Ye, J.; Fang, L.; Zheng, H.K.; Zhang, Y.; Chen, J.; Zhang, Z.; Wang, J.; Li, S.; Li, R.; Bolund, L. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006, 34, 293–297. [Google Scholar] [CrossRef]
- Li, R.; Zhang, C.L.; Liao, X.X.; Chen, D.; Wang, W.Q.; Zhu, Y.H.; Geng, X.H.; Ji, D.J.; Mao, Y.J.; Gong, Y.C. Transcriptome microrna profiling of bovine mammary glands infected with staphylococcus aureus. Int. J. Mol. Sci. 2015, 16, 4997–5013. [Google Scholar] [CrossRef] [Green Version]
- Ernst, J.; Joseph, Z.B. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinform. 2006, 7, 191. [Google Scholar] [CrossRef] [Green Version]
- Saldanha, A.J. Java Treeview--extensible visualization of microarray data. Bioinformatics 2004, 20, 3246–3248. [Google Scholar] [CrossRef] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101. [Google Scholar] [CrossRef]
- Ye, W.; Wu, H.Q.; He, X.; Wang, L.; Zhang, W.M.; Li, H.H.; Fan, Y.F.; Tan, G.H.; Liu, T.; Gao, X.X. Transcriptome sequencing of chemically induced Aquilaria sinensis to identify genes related to agarwood formation. PLoS ONE 2016, 11, e0155505. [Google Scholar] [CrossRef] [PubMed]









© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, W.; Liu, T.; Zhang, W.; Li, S.; Zhu, M.; Li, H.; Kong, Y.; Xu, L. Disclosure of the Molecular Mechanism of Wheat Leaf Spot Disease Caused by Bipolaris sorokiniana through Comparative Transcriptome and Metabolomics Analysis. Int. J. Mol. Sci. 2019, 20, 6090. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20236090
Ye W, Liu T, Zhang W, Li S, Zhu M, Li H, Kong Y, Xu L. Disclosure of the Molecular Mechanism of Wheat Leaf Spot Disease Caused by Bipolaris sorokiniana through Comparative Transcriptome and Metabolomics Analysis. International Journal of Molecular Sciences. 2019; 20(23):6090. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20236090
Chicago/Turabian StyleYe, Wei, Taomei Liu, Weimin Zhang, Saini Li, Muzi Zhu, Haohua Li, Yali Kong, and Liqiong Xu. 2019. "Disclosure of the Molecular Mechanism of Wheat Leaf Spot Disease Caused by Bipolaris sorokiniana through Comparative Transcriptome and Metabolomics Analysis" International Journal of Molecular Sciences 20, no. 23: 6090. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20236090