Effect of Trichoderma velutinum and Rhizoctonia solani on the Metabolome of Bean Plants (Phaseolus vulgaris L.)
Abstract
:1. Introduction
2. Results
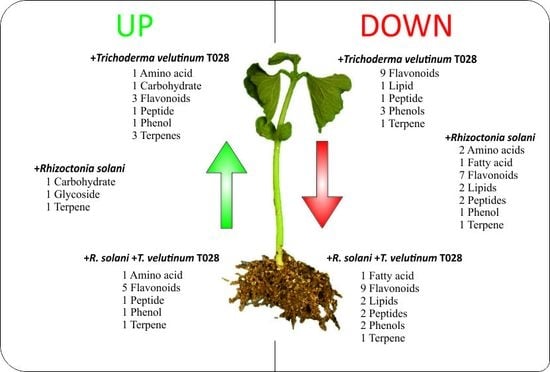
2.1. Analysis of Phaseolus vulgaris Plant Metabolome in Presence of T. velutinum and R. solani
2.2. Characterization of Compounds in Bean Leaf Metabolome
2.3. Amino Acids and Peptides
2.4. Carbohydrates and Glycosides
2.5. Fatty Acids and Lipids
2.6. Phenols
2.7. Terpenes
2.8. Principal Component Analysis (PCA)
3. Discussion
4. Material and Methods
4.1. Fungal Isolates and Culture Conditions
4.2. Plant Material and Growth Conditions
4.3. Preparation of Bean Leaf Extracts
4.4. LC-MS qTOF Conditions
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Guerrero-González, M.L.; Rodríguez-Kessler, M.; Rodríguez-Guerra, R.; González-Chavira, M.; Simpson, J.; Sánchez, F.; Jiménez-Bremont, J.F. Differential expression of Phaseolus vulgaris genes induced during the interaction with Rhizoctonia solani. Plant Cell Rep. 2011, 30, 1465–1473. [Google Scholar] [CrossRef] [PubMed]
- Pieterse, C.M.J.; Leon-Reyes, A.; Van der Ent, S.; Van Wees, S.C.M. Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 2009, 5, 308–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denancé, N.; Sánchez-Vallet, A.; Goffner, D.; Molina, A. Disease resistance or growth: The role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 2013, 4, 155. [Google Scholar] [CrossRef] [PubMed]
- Mhlongo, M.I.; Steenkamp, P.A.; Piater, L.A.; Madala, N.E.; Dubery, I.A. Profiling of altered metabolomic states in Nicotiana tabacum cells induced by priming agents. Front. Plant Sci. 2016, 7, 1527. [Google Scholar] [CrossRef] [PubMed]
- Hagedorn, D.J. Rhizoctonia Root Rot. In Compendium of Bean Diseases; Hall, R., Ed.; APS, American Phytopathological Society: Saint Paul, MN, USA, 1991; p. 13. [Google Scholar]
- Hermosa, R.; Belén Rubio, M.; Cardoza, R.E.; Nicolás, C.; Monte, E.; Gutiérrez, S. The contribution of Trichoderma to balancing the costs of plant growth and defense. Int. Microbiol. 2013, 16, 69–80. [Google Scholar]
- Shoresh, M.; Harman, G.E.; Mastouri, F. Induced systemic resistance and plant responses to fungal biocontrol agents. Annu. Rev. Phytopathol. 2010, 48, 21–43. [Google Scholar] [CrossRef] [PubMed]
- Mayo, S.; Cominelli, E.; Sparvoli, F.; González-López, O.; Rodríguez-González, A.; Gutiérrez, S.; Casquero, P.A. Development of a qPCR strategy to select bean genes involved in plant defense response and regulated by the Trichoderma velutinum–Rhizoctonia solani interaction. Front. Plant Sci. 2016, 7, 1109. [Google Scholar] [CrossRef] [PubMed]
- Heuberger, A.L.; Robison, F.M.; Lyons, S.M.A.; Broeckling, C.D.; Prenni, J.E. Evaluating plant immunity using mass spectrometry-based metabolomics workflows. Front. Plant Sci. 2014, 5, 291. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Qi, X.; Duan, L.-X. Overview. In Plant Metabolomics; Qi, X., Chen, X., Wang, Y., Eds.; Springer: Dordrecht, The Netherlands, 2015; pp. 1–24. [Google Scholar]
- Abu-Reidah, I.M.; Arráez-Román, D.; Lozano-Sánchez, J.; Segura-Carretero, A.; Fernández-Gutiérrez, A. Phytochemical characterisation of green beans (Phaseolus vulgaris L.) by using high-performance liquid chromatography coupled with time-of-flight mass spectrometry. Phytochem. Anal. 2013, 24, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.; Mathesius, U. The role of flavonoids in root-rhizosphere signalling: Opportunities and challenges for improving plant-microbe interactions. J. Exp. Bot. 2012, 63, 3429–3444. [Google Scholar] [CrossRef] [PubMed]
- Harrison, M.J.; Dixon, R.A. Spatial patterns of expression of flavonoid/isoflavonoid pathway genes during interactions between roots of Medicago truncatula and the mycorrhizal fungus Glomus versiforme. Plant J. 1994, 6, 9–20. [Google Scholar] [CrossRef]
- Taiz, L.; Zeiger, E.; Møller, I.M.; Murphy, A.S. Plant Physiology and Development, 6th ed.; Taiz, L., Møller, I.M., Murphy, A., Zeiger, E., Eds.; Sinauer Associates, Incorporated: Sunderland, MA, USA, 2015; ISBN 9781605352558. [Google Scholar]
- Wink, M. Evolution of secondary metabolites from an ecological and molecular phylogenetic perspective. Phytochemistry 2003, 64, 3–19. [Google Scholar] [CrossRef]
- Wang, W.; Zhou, H.; Lin, H.; Roy, S.; Shaler, T.A.; Hill, L.R.; Norton, S.; Kumar, P.; Anderle, M.; Becker, C.H. Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Anal. Chem. 2003, 75, 4818–4826. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Lam, H.; Pi, E.; Zhan, Q.; Tsai, S.; Wang, C.; Kwan, Y.; Ngai, S. Comparative metabolomics in Glycine max and Glycine soja under salt stress to reveal the phenotypes of their offspring. J. Agric. Food Chem. 2013, 61, 8711–8721. [Google Scholar] [CrossRef] [PubMed]
- Brechenmacher, L.; Lei, Z.; Libault, M.; Findley, S.; Sugawara, M.; Sadowsky, M.J.; Sumner, L.W.; Stacey, G. Soybean metabolites regulated in root hairs in response to the symbiotic bacterium Bradyrhizobium japonicum. Plant Physiol. 2010, 153, 1808–1822. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa Laboratories KEGG PATHWAY: Alpha-Linolenic Acid Metabolism—Phaseolus Vulgaris (Common Bean). Available online: http://www.kegg.jp/kegg-bin/show_pathway?org_name=pvu&mapno=00592&mapscale=&show_description=hide (accessed on 14 April 2018).
- Yannai, S. Dictionary of Food Compounds with CD-ROM, 2nd ed.; Yannai, S., Ed.; CRC Press Taylor & Francis Group: Boca Raton, FL, USA, 2012; ISBN 13:978-1-4200-8352-1. [Google Scholar]
- Buckingham, J.; Munasinghe, V.R.N. Dictionary of Flavonoids; Buckingham, J., Munasinghe, V.R.N., Eds.; Taylor & Francis Group: Boca Raton, FL, USA, 2015; ISBN 9781466554344. [Google Scholar]
- Wojakowska, A.; Muth, D.; Narożna, D.; Mądrzak, C.; Stobiecki, M.; Kachlicki, P. Changes of phenolic secondary metabolite profiles in the reaction of narrow leaf lupin (Lupinus angustifolius) plants to infections with Colletotrichum lupini fungus or treatment with its toxin. Metabolomics 2013, 9, 575–589. [Google Scholar] [CrossRef] [PubMed]
- Wojakowska, A.; Kułak, K.; Jasiński, M.; Kachlicki, P.; Stawiński, S.; Stobiecki, M. Metabolic response of narrow leaf lupine (Lupinus angustifolius) plants to elicitation and infection with Colletotrichum lupini under field conditions. Acta Physiol. Plant. 2015, 37, 152. [Google Scholar] [CrossRef]
- Southon, I.W.; Bisby, F.A.; Buckingham, J.; Harborne, J.B.; Zarucchi, J.L. Phytochemical Dictionary of the Leguminosae, 1st ed.; Bisby, F.A., Buckingham, J., Harbome, J.B., Eds.; Springer-Science + Business Media: London, UK, 1994; ISBN 978-0-412-39770-7. [Google Scholar]
- Rizk, A.-F.; Wood, G.E.; Salunkhe, D.K. Phytoalexins of leguminous plants. CRC Crit. Rev. Food Sci. Nutr. 1980, 13, 245–295. [Google Scholar] [CrossRef]
- Pushpa, D.; Yogendra, K.N.; Gunnaiah, R.; Kushalappa, A.C.; Murphy, A. Identification of late blight resistance-related metabolites and genes in potato through nontargeted metabolomics. Plant Mol. Biol. Rep. 2014, 32, 584–595. [Google Scholar] [CrossRef]
- Contreras-Cornejo, H.A.; Macías-Rodríguez, L.; López-Bucio, J.S.; López-Bucio, J. Enhanced plant immunity using Trichoderma. In Biotechnology and Biology of Trichoderma; Gupta, V.G., Schmoll, M., Herrera-Estrella, A., Upadhyay, R.S., Druzhinina, I., Tuohy, M., Eds.; Elsevier: Oxford, UK, 2014; pp. 495–504. ISBN 9780444595768. [Google Scholar]
- Robison, F.M.; Turner, M.F.; Jahn, C.E.; Schwartz, H.F.; Prenni, J.E.; Brick, M.A.; Heuberger, A.L. Common bean varieties demonstrate differential physiological and metabolic responses to the pathogenic fungus Sclerotinia sclerotiorum. Plant Cell Environ. 2018, 41, 2141–2154. [Google Scholar] [CrossRef]
- VanEtten, H.D.; Mansfield, J.W.; Bailey, J.A.; Farmer, E.E. Two Classes of Plant Antibiotics: Phytoalexins versus “Phytoanticipins”. Plant Cell 1994, 6, 1191–1192. [Google Scholar] [CrossRef] [PubMed]
- Ahuja, I.; Kissen, R.; Bones, A.M. Phytoalexins in defense against pathogens. Trends Plant Sci. 2012, 17, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, J.P.; Osbourn, A.E. Fungal resistance to plant antibiotics as a mechanism of pathogenesis. Microbiol. Mol. Biol. Rev. 1999, 63, 708–724. [Google Scholar] [PubMed]
- Hammerschmidt, R.; Becker, J.S. Acquired resistance to disease in plants. In Horticultural Reviews; Janick, J., Ed.; John Wiley & Sons: New York, NY, USA, 1997; Volume 18, pp. 247–280. ISBN 0-471-57334-5. [Google Scholar]
- Pieterse, C.M.J.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C.M. Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 2012, 28, 489–521. [Google Scholar] [CrossRef] [PubMed]
- Glawischnig, E. Camalexin. Phytochemistry 2007, 68, 401–406. [Google Scholar] [CrossRef] [PubMed]
- Pourcel, L.; Routaboul, J.; Cheynier, V.; Lepiniec, L.; Debeaujon, I. Flavonoid oxidation in plants: From biochemical properties to physiological functions. Trends Plant Sci. 2007, 12, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Kuc, J. Phytoalexins, stress metabolism, and disease resistance in plants. Annu. Rev. Phytopathol. 1991, 33, 275–297. [Google Scholar] [CrossRef]
- Yogendra, K.N.; Kushalappa, A.C.; Sarmiento, F.; Rodriguez, E.; Mosquera, T. Metabolomics deciphers quantitative resistance mechanisms in diploid potato clones against late blight. Funct. Plant Biol. 2015, 42, 284–298. [Google Scholar] [CrossRef]
- Heitz, T.; Smirnova, E.; Widemann, E.; Aubert, Y.; Pinot, F.; Ménard, R. The rise and fall of jasmonate biological activities. In Lipids in Plant and Algae Development; Springer International Publishing: New York, NY, USA, 2016; pp. 405–426. [Google Scholar]
- Druzhinina, I.S.; Seidl-Seiboth, V.; Herrera-Estrella, A.; Horwitz, B.A.; Kenerley, C.M.; Monte, E.; Mukherjee, P.K.; Zeilinger, S.; Grigoriev, I.V.; Kubicek, C.P. Trichoderma: The genomics of opportunistic success. Nat. Rev. Microbiol. 2011, 9, 749–759. [Google Scholar] [CrossRef]
- Vinale, F.; Sivasithamparam, K.; Ghisalberti, E.L.; Ruocco, M.; Woo, S.; Lorito, M. Trichoderma secondary metabolites that affect plant metabolism. Nat. Prod. Commun. 2012, 7, 1545–1550. [Google Scholar]
- Hoyos-Carvajal, L.; Orduz, S.; Bissett, J. Growth stimulation in bean (Phaseolus vulgaris L.) by Trichoderma. Biol. Control 2009, 51, 409–416. [Google Scholar] [CrossRef]
- Qureshi, M.A.; Iqbal, A.; Akhtar, N.; Shakir, M.A.; Khan, A. Co-inoculation of phosphate solubilizing bacteria and rhizobia in the presence of L-tryptophan for the promotion of mash bean (Vigna mungo L.). Soil Environ. 2012, 31, 47–54. [Google Scholar]
- Luna-Vital, D.A.; Mojica, L.; González de Mejía, E.; Mendoza, S.; Loarca-Piña, G. Biological potential of protein hydrolysates and peptides from common bean (Phaseolus vulgaris L.): A review. Food Res. Int. 2015, 76, 39–50. [Google Scholar] [CrossRef]
- Mayo, S.; Gutierrez, S.; Malmierca, M.G.; Lorenzana, A.; Campelo, M.P.; Hermosa, R.; Casquero, P.A. Influence of Rhizoctonia solani and Trichoderma spp. in growth of bean (Phaseolus vulgaris L.) and in the induction of plant defense-related genes. Front. Plant Sci. 2015, 6, 685. [Google Scholar] [CrossRef] [PubMed]
- Talhaoui, N.; Gómez-Caravaca, A.M.; Roldán, C.; León, L.; De la Rosa, R.; Fernández-Gutiérrez, A.; Segura-Carretero, A. Chemometric analysis for the evaluation of phenolic patterns in olive leaves from six cultivars at different growth stages. J. Agric. Food Chem. 2015, 63, 1722–1729. [Google Scholar] [CrossRef]




| Peak | Experimental Mass | m/z | Score | Diff (ppm) | RT (min) | Formula | Putative Identification | Reference |
|---|---|---|---|---|---|---|---|---|
| Amino acids | ||||||||
| 8 | 204.0898 | 203.0826 [M − H]− 407.1733 [2M − H]− | 49.94 | −0.09 | 8.52 | C11H12N2O2 | l-Tryptophan | PubChem ID number—6305 [17] |
| 15 | 246.1004 | 245.09315 [M − H]− | 99.91 | 0.05 | 14.20 | C13H14N2O3 | N-acetyltryptophan | PubChem ID number—2002 [17] |
| Peptides | ||||||||
| 7 | 230.1629 | 229.1557 [M − H]− | 87.05 | 0.28 | 8.42 | C11H22N2O3 | Valyl-Leucine | PubChem ID number—107487 |
| 11 | 372.2371 | 371.2298 [M − H]− 743.4673 [2M − H]− | 99.84 | 0.41 | 10.00 | C17H32N4O5 | Ile Gln Ile | NA |
| 29 | 402.1863 | 401.1788 [M − H]− | 98.04 | 0.70 | 19.94 | C15H26N6O7 | Gln Gln Gln | NA |
| Carbohydrates | ||||||||
| 2 | 342.1159 | 341.1087 [M − H]− | 99.71 | 0.83 | 1.43 | C12H22O11 | Sucrose | PubChem ID number—5988 [17] |
| Glycosides | ||||||||
| 4 | 338.0848 | 337.0775 [M − H]− | 85.70 | 0.87 | 4.96 | C12H18O11 | l-Ascorbic acid-2-glucoside | PubChem ID number—54693473 |
| Fatty acids | ||||||||
| 35 | 278.22338 | 277.2169 [M − H]− 555.4410 [2M − H]− | 84.78 | 2.26 | 30.59 | C18H30O2 | γ-Linolenic acid | PubChem ID number—5280933 [17,18,19] |
| Lipids | ||||||||
| Fatty acyls—Octadecanoids | ||||||||
| 34 | 294.2195 | 293.2122 [M − H]− | 99.67 | 0.76 | 26.29 | C18H30O3 | 13(S)-HOTrE | PubChem ID number—47205624 |
| Glycerophosphates | ||||||||
| 36 | 578.4296 | 577.4226 [M − H]− | 90.87 | 0.43 | 30.59 | C31H63O7P | PA(O-16:0/12:0) | PubChem number—52929565 |
| Fatty acyls glycoside | ||||||||
| 30 | 334.1990 | 333.1916 [M − H]− | 89.56 | 0.38 | 19.94 | C16H30O7 | 3-O-α-l-rhamnopyranosyl-3-hydroxydecanoic acid | PubChem ID number—56936287 [20,21] |
| Flavonoids | ||||||||
| Flavone | ||||||||
| 22 | 374.0998 | 373.0926 [M − H]− | 99.35 | 0.86 | 16.90 | C19H18O8 | 3′,5-Dihydroxy-3,4′,6,7-tetramethoxyflavone | [21] |
| 27 | 390.0951 | 389.0874 [M − H]− | 82.88 | 1.10 | 18.32 | C19H18O9 | 5,2′,4′-Trihydroxy-3,7,8,5′-tetramethoxyflavone | PubChem ID number—85296959 |
| Flavonol | ||||||||
| 12 | 596.1739 | 595.1666 [M − H]− | 99.67 | 0.33 | 10.27 | C26H28O16 | Quercetin 3-vicianoside | PubChem Id number—44259139 [11] |
| 26 | 454.2409 | 453.2338 [M − H]− | 99.76 | 0.29 | 18.05 | C20H38O11 | n-Octyl-β-d-maltopyranoside | NA |
| Flavonol | ||||||||
| 21 | 414.2248 | 413.2177 [M − H]− | 82.59 | 0.37 | 16.25 | C29H50O | Sitosterol | PubChem ID number—86821 [17,21] |
| Flavonone | ||||||||
| 3 | 580.1847 | 579.1773 [M − H]− | 99.42 | 0.72 | 1.47 | C27H32O14 | Naringin | PubChem ID number—442428 [17] |
| Flavonoids glycoconjugate | ||||||||
| 1 | 400.1006 | 399.0930 [M − H]− | 49.90 | 0.30 | 1.31 | C17H20O11 | 5-Hydroxy-6,8-dimethoxy-2-oxo-2H-chromen-7-yl β-d-glucopyranoside | NA |
| 10 | 612.1686 | 611.1613 [M − H]− | 99.03 | 0.75 | 9.37 | C27H32O16 | 3,4′,5,7-Tetrahydroxyflavanone 3,7-Di-O-β-d-glucopyranoside | CAS Number—80212-10-8 [21] |
| 14 | 552.1841 | 551.1762 [M − H]− | 97.55 | 1.60 | 11.13 | C26H32O13 | (Z)-Resveratrol 3,4′-diglucoside | PubChem ID number—22298557 [20] |
| 16 | 642.1193 | 641.1119 [M − H]− | 99.69 | 0.09 | 14.34 | C30H26O16 | Quercetagetin 7-(6″-(E)-caffeoylglucoside) | PubChem Id number—44259848 |
| 18 | 654.1769 | 653.1697 [M − H]− | 99.89 | −0.20 | 16.11 | C29H34O17 | Isopyrenin 7-O-glucoside | CAS Number—61252-86-6 [21] |
| 19 | 540.1838 | 539.1766 [M − H]− | 99.49 | 0.89 | 16.11 | C25H32O13 | 12-Hydroxy, O-[3,4,5-trihydroxybenzoyl-(06)-β-d-glucopyranoside] | [20] |
| 20 | 678.2894 | 677.2822 [M − H]− | 85.36 | 0.98 | 16.24 | C32H39O16 | Luteone 4,7-O-diglucoside | [22,23] |
| 24 | 470.2359 | 469.2287 [M − H]− | 98.56 | 0.25 | 17.61 | C20H38O12 | (R)-1-O-[β-d-Glucopyranosyl-(1-6)-β-d-glucopyranoside]-1,3-octanediol | HMDB Id number—32799 |
| Isoflavanoid | ||||||||
| 28 | 338.1522 | 337.1451[M − H]− 319.1298 [M − H2O]− 168.0663 [M − 2H]− | 78.85 | 0.31 | 8.38 | C21H22O4 | 2′-O-Methylphaseollinisoflavan | CAS Number—49594-01-6 [19,23,24] |
| Isoflavanone | ||||||||
| 25 | 424.2304 | 423.2234 [M − H]− | 98.58 | 0.58 | 17.62 | C25H28O6 | 2′,4′,5,7-Tetrahydroxy-3′,8-diprenylisoflavanone | CAS Number—64280-18-8 [24] |
| Isoflavans | ||||||||
| 6 | 324.1419 | 323.1346 [M − H]− 647.2765 [2M − H]− | 99.89 | 0.29 | 8.13 | C20H20O4 | Phaseollinisoflavan | PubChem ID number—4484952 [25] |
| Phenols | ||||||||
| Tyrosols | ||||||||
| 5 | 316.1153 | 315.1082 [M − H]− | 97.00 | 1.57 | 7.29 | C14H20O8 | Hydroxytyrosol 1-O-glucoside | PubChem ID number—13845930 |
| Phenylpropanoids | ||||||||
| 13 | 386.1201 | 385.1127 [M − H]− | 94.90 | 3.17 | 10.71 | C17H22O10 | 1-O-Sinapoylglucose | PubChem ID number—5280406 |
| Xanthonoid | ||||||||
| 31 | 392.1102 | 391.1032 [M − H]− | 97.71 | 1.07 | 21.10 | C19H20O9 | Garcimangosone D | PubChem ID number—11003703 [20] |
| 33 | 628.3063 | 627.2991 [M − H]− | 99.44 | 0.83 | 25.77 | C38H44O8 | Gambogic acid | PubChem ID number—5281632 |
| Terpenes | ||||||||
| Terpenoid | ||||||||
| 9 | 444.1995 | 443.1923 [M − H]− | 99.84 | 0.06 | 8.88 | C21H32O10 | Dihydrophaseic acid 4-O-β-d-glucoside | ChEBI Id number—23758 [26] |
| Triterpenoid | ||||||||
| 32 | 896.5127 | 895.5050 [M − H]− | 98.64 | 0.47 | 24.97 | C47H76O16 | Akeboside Ste | PubChem ID number—46173935 |
| Terpene glycosides | ||||||||
| 17 | 416.2046 | 415.1971 [M − H]− | 99.53 | 0.52 | 14.78 | C20H32O9 | Ethyl 7-epi-12-hydroxyjasmonate glucoside | HMDB Id number—36340 [17] |
| Terpene | ||||||||
| 23 | 268.1308 | 267.1235 [M − H]− 535.2548 [2M − H]− | 49.76 | 1.09 | 17.52 | C14H20O5 | Teucrein | ChemSpider ID number—28944862 |
| Id | Formula | Putative Identification | CT028 vs. CC a | CR vs. CC a | RT028 vs. CC a |
|---|---|---|---|---|---|
| Amino acids | |||||
| 8 | C11H12N2O2 | l-Tryptophan | Up | Down | -- |
| 15 | C13H14N2O3 | N-acetyltryptophan | -- | Down | Up |
| Peptides | |||||
| 7 | C11H22N2O3 | Valyl-Leucine | Up | Down | Up |
| 11 | C17H32N4O5 | Ile Gln Ile | -- | Down | Down |
| 29 | C15H26N6O7 | Gln Gln Gln | Down | -- | Down |
| Carbohydrates | |||||
| 2 | C12H22O11 | Sucrose | Up | Up | -- |
| Glycosides | |||||
| 4 | C12H18O11 | l-Ascorbic acid-2-glucoside | -- | Up | -- |
| Fatty acids | |||||
| 35 | C18H30O2 | γ-Linolenic acid | -- | Down | Down |
| Lipids | |||||
| 30 | C16H30O7 | 3-O-α-l-rhamnopyranosyl-3-hydroxydecanoic acid | Down | -- | Down |
| 34 | C18H30O3 | 13(S)-HOTrE | -- | Down | -- |
| 36 | C31H63O7P | PA(O-16:0/12:0) | -- | Down | Down |
| Flavonoids | |||||
| 1 | C17H20O11 | 5-Hydroxy-6,8-dimethoxy-2-oxo-2H-chromen-7-yl β-d-glucopyranoside | Down | Down | Down |
| 3 | C27H32O14 | Naringin | Up | -- | -- |
| 6 | C20H20O4 | Phaseollinisoflavan | -- | -- | Down |
| 10 | C27H32O16 | 3,4’,5,7-Tetrahydroxyflavanone 3,7-Di-O-β-d-glucopyranoside | Up | -- | Up |
| 12 | C26H28O16 | Quercetin 3-vicianoside | -- | -- | Up |
| 14 | C26H32O13 | (Z)-Resveratrol 3,4′-diglucoside | -- | -- | Up |
| 16 | C30H26O16 | Quercetagetin 7-(6″-(E)-caffeoylglucoside) | -- | Down | -- |
| 18 | C29H34O17 | Isopyrenin 7-O-glucoside | Down | Down | Down |
| 19 | C25H32O13 | 12-Hydroxy, O-[3,4,5-trihydroxybenzoyl-(06)-β-d-glucopyranoside] | Down | Down | Down |
| 20 | C32H39O16 | Luteone 4,7-O-diglucoside | Up | Down | Up |
| 21 | C29H50O | Sitosterol | -- | -- | Up |
| 22 | C19H18O8 | 3′,5-Dihydroxy-3,4′,6,7-tetramethoxyflavone | Down | Down | Down |
| 24 | C20H38O12 | (R)-1-O-[β-d-Glucopyranosyl-(1-6)-β-d-glucopyranoside]-1,3-octanediol | Down | -- | Down |
| 25 | C25H28O6 | 2′,4′,5,7-Tetrahydroxy-3′,8-diprenylisoflavanone | Down | -- | Down |
| 26 | C20H38O11 | n-Octyl-β-d-maltopyranoside | Down | -- | -- |
| 27 | C19 H18 O9 | 5,2’,4′-Trihydroxy-3,7,8,5′-tetramethoxyflavone | Down | Down | Down |
| 28 | C21H22O4 | 2′-O-Methylphaseollinisoflavan | Down | -- | Down |
| Phenols | |||||
| 5 | C14H20O8 | Hydroxytyrosol 1-O-glucoside | Down | -- | -- |
| 13 | C17H22O10 | 1-O-Sinapoylglucose | Up | -- | Up |
| 31 | C19H20O9 | Garcimangosone D | Down | Down | Down |
| 33 | C38H44O8 | Gambogic acid | Down | -- | Down |
| Terpenes | |||||
| 9 | C21H32O10 | Dihydrophaseic acid 4-O-β-d-glucoside | Up | -- | -- |
| 17 | C20H32O9 | Ethyl 7-epi-12-hydroxyjasmonate glucoside | Up | Up | Up |
| 23 | C14H20O5 | Teucrein | Up | -- | -- |
| 32 | C47H76O16 | Akeboside Ste | Down | Down | Down |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mayo-Prieto, S.; Marra, R.; Vinale, F.; Rodríguez-González, Á.; Woo, S.L.; Lorito, M.; Gutiérrez, S.; Casquero, P.A. Effect of Trichoderma velutinum and Rhizoctonia solani on the Metabolome of Bean Plants (Phaseolus vulgaris L.). Int. J. Mol. Sci. 2019, 20, 549. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20030549
Mayo-Prieto S, Marra R, Vinale F, Rodríguez-González Á, Woo SL, Lorito M, Gutiérrez S, Casquero PA. Effect of Trichoderma velutinum and Rhizoctonia solani on the Metabolome of Bean Plants (Phaseolus vulgaris L.). International Journal of Molecular Sciences. 2019; 20(3):549. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20030549
Chicago/Turabian StyleMayo-Prieto, Sara, Roberta Marra, Francesco Vinale, Álvaro Rodríguez-González, Sheridan L. Woo, Matteo Lorito, Santiago Gutiérrez, and Pedro A. Casquero. 2019. "Effect of Trichoderma velutinum and Rhizoctonia solani on the Metabolome of Bean Plants (Phaseolus vulgaris L.)" International Journal of Molecular Sciences 20, no. 3: 549. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20030549