Transcriptional Analysis of Immunohistochemically Defined Subgroups of Non-Muscle-Invasive Papillary High-Grade Upper Tract Urothelial Carcinoma
Abstract
:1. Introduction
2. Results
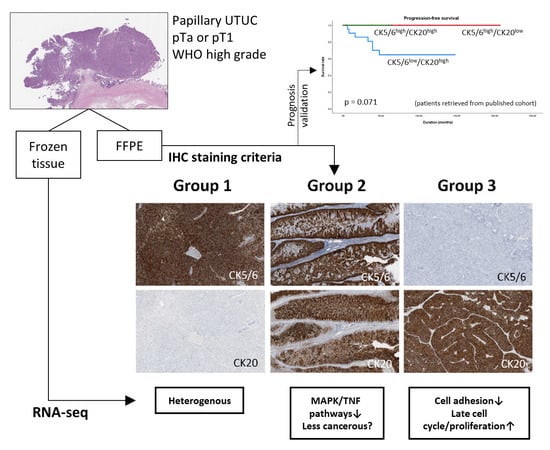
2.1. Subgroups of Non-Muscle-Invasive High-Grade Papillary UTUC Based on IHC Staining for CK5/6 and CK20
2.2. Identification of Differentially Expressed Genes (DEGs) between the Subgroups
2.3. Functional Analysis of the Subgroups
2.3.1. Group 3 was Enriched for Processes Related to Cellular Adhesion and Motility
2.3.2. Group 2 Showed Downregulation of Mitogen-Activated Protein Kinase (MAPK) and Tumor Necrosis Factor (TNF) Signaling Pathways
2.3.3. Expression of Biologic Signature Genes of the Subgroups
3. Discussion
4. Materials and Methods
4.1. Sample Selection and Immunohistochemistry
4.2. Validation of Prognosis Associated with Immunohistochemical Profiles of RNA-Seq Subgroups
4.3. Transcription Analysis and Identification of Differentially Expressed Genes
4.4. Functional Enrichment Analysis
4.5. TCGA Data Preprocessing and Survival Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| MIBC | muscle-invasive bladder cancer |
| NMIBC | non-muscle-invasive bladder cancer |
| CIS | carcinoma in situ |
| UTUC | upper tract urothelial carcinoma |
| IHC | immunohistochemistry |
| BASQ | basal/squamous-like |
| RNA-seq | RNA sequencing |
| PFS | progression-free survival |
| DEG | differentially expressed gene |
| FDR | false discovery rate |
| GO | gene ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| BP | biologic process |
| CC | cellular component |
| MF | molecular function |
| GSEA | gene set enrichment analysis |
| TCGA | The Cancer Genome Atlas |
| MAPK | mitogen-activated protein kinase |
| TNF | tumor necrosis factor |
References
- Sjodahl, G.; Lauss, M.; Lovgren, K.; Chebil, G.; Gudjonsson, S.; Veerla, S.; Patschan, O.; Aine, M.; Ferno, M.; Ringner, M.; et al. A molecular taxonomy for urothelial carcinoma. Clin. Cancer. Res. 2012, 18, 3377–3386. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.; Porten, S.; Kim, S.; Willis, D.; Plimack, E.R.; Hoffman-Censits, J.; Roth, B.; Cheng, T.; Tran, M.; Lee, I.L.; et al. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell 2014, 25, 152–165. [Google Scholar] [CrossRef] [PubMed]
- Damrauer, J.S.; Hoadley, K.A.; Chism, D.D.; Fan, C.; Tiganelli, C.J.; Wobker, S.E.; Yeh, J.J.; Milowsky, M.I.; Iyer, G.; Parker, J.S.; et al. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc. Natl. Acad. Sci. USA 2014, 111, 3110–3115. [Google Scholar] [CrossRef] [Green Version]
- The Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014, 507, 315–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, A.G.; Kim, J.; Al-Ahmadie, H.; Bellmunt, J.; Guo, G.; Cherniack, A.D.; Hinoue, T.; Laird, P.W.; Hoadley, K.A.; Akbani, R.; et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 2017, 171, 540–556.e25. [Google Scholar] [CrossRef] [PubMed]
- Hedegaard, J.; Lamy, P.; Nordentoft, I.; Algaba, F.; Hoyer, S.; Ulhoi, B.P.; Vang, S.; Reinert, T.; Hermann, G.G.; Mogensen, K.; et al. Comprehensive transcriptional analysis of early-stage urothelial carcinoma. Cancer Cell 2016, 30, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Aine, M.; Eriksson, P.; Liedberg, F.; Sjodahl, G.; Hoglund, M. Biological determinants of bladder cancer gene expression subtypes. Sci. Rep. 2015, 5, 10957. [Google Scholar] [CrossRef] [Green Version]
- Seiler, R.; Ashab, H.A.D.; Erho, N.; van Rhijn, B.W.G.; Winters, B.; Douglas, J.; Van Kessel, K.E.; Fransen van de Putte, E.E.; Sommerlad, M.; Wang, N.Q.; et al. Impact of molecular subtypes in muscle-invasive bladder cancer on predicting response and survival after neoadjuvant chemotherapy. Eur. Urol. 2017, 72, 544–554. [Google Scholar] [CrossRef]
- McConkey, D.J.; Choi, W.; Ochoa, A.; Siefker-Radtke, A.; Czerniak, B.; Dinney, C.P. Therapeutic opportunities in the intrinsic subtypes of muscle-invasive bladder cancer. Hematol. Oncol. Clin. N. Am. 2015, 29, 377–394. [Google Scholar] [CrossRef]
- McConkey, D.J.; Choi, W.; Shen, Y.; Lee, I.L.; Porten, S.; Matin, S.F.; Kamat, A.M.; Corn, P.; Millikan, R.E.; Dinney, C.; et al. A prognostic gene expression signature in the molecular classification of chemotherapy-naive urothelial cancer is predictive of clinical outcomes from neoadjuvant chemotherapy: A phase 2 trial of dose-dense methotrexate, vinblastine, doxorubicin, and cisplatin with bevacizumab in urothelial cancer. Eur. Urol. 2016, 69, 855–862. [Google Scholar] [CrossRef]
- Hurst, C.D.; Alder, O.; Platt, F.M.; Droop, A.; Stead, L.F.; Burns, J.E.; Burghel, G.J.; Jain, S.; Klimczak, L.J.; Lindsay, H.; et al. Genomic subtypes of non-invasive bladder cancer with distinct metabolic profile and female gender bias in KDM6A mutation frequency. Cancer Cell 2017, 32, 701–715.e07. [Google Scholar] [CrossRef] [PubMed]
- Roupret, M.; Babjuk, M.; Comperat, E.; Zigeuner, R.; Sylvester, R.J.; Burger, M.; Cowan, N.C.; Gontero, P.; Van Rhijn, B.W.G.; Mostafid, A.H.; et al. European association of urology guidelines on upper urinary tract urothelial carcinoma: 2017 update. Eur. Urol. 2018, 73, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Sfakianos, J.P.; Cha, E.K.; Iyer, G.; Scott, S.N.; Zabor, E.C.; Shah, R.H.; Ren, Q.; Bagrodia, A.; Kim, P.H.; Hakimi, A.A.; et al. Genomic characterization of upper tract urothelial carcinoma. Eur. Urol. 2015, 68, 970–977. [Google Scholar] [CrossRef]
- Sanford, T.; Porten, S.; Meng, M.V. Molecular analysis of upper tract and bladder urothelial carcinoma: Results from a microarray comparison. PLoS ONE 2015, 10, e0137141. [Google Scholar] [CrossRef] [PubMed]
- Glaser, A.P.; Fantini, D.; Shilatifard, A.; Schaeffer, E.M.; Meeks, J.J. The evolving genomic landscape of urothelial carcinoma. Nat. Rev. Urol. 2017, 14, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Audenet, F.; Attalla, K.; Sfakianos, J.P. The evolution of bladder cancer genomics: What have we learned and how can we use it? Urol. Oncol. 2018, 36, 313–320. [Google Scholar] [CrossRef] [PubMed]
- Dadhania, V.; Zhang, M.; Zhang, L.; Bondaruk, J.; Majewski, T.; Siefker-Radtke, A.; Guo, C.C.; Dinney, C.; Cogdell, D.E.; Zhang, S.; et al. Meta-analysis of the luminal and basal subtypes of bladder cancer and the identification of signature immunohistochemical markers for clinical use. EBioMedicine 2016, 12, 105–117. [Google Scholar] [CrossRef]
- Lerner, S.P.; McConkey, D.J.; Hoadley, K.A.; Chan, K.S.; Kim, W.Y.; Radvanyi, F.; Hoglund, M.; Real, F.X. Bladder cancer molecular taxonomy: Summary from a consensus meeting. Bladder Cancer 2016, 2, 37–47. [Google Scholar] [CrossRef]
- Sikic, D.; Keck, B.; Wach, S.; Taubert, H.; Wullich, B.; Goebell, P.J.; Kahlmeyer, A.; Olbert, P.; Isfort, P.; Nimphius, W.; et al. Immunohistochemiocal subtyping using CK20 and CK5 can identify urothelial carcinomas of the upper urinary tract with a poor prognosis. PLoS ONE 2017, 12, e0179602. [Google Scholar] [CrossRef]
- Jung, M.; Kim, B.; Moon, K.C. Immunohistochemistry of CK5/6, CD44, and CK20 as prognostic biomarkers of non-muscle-invasive papillary upper tract urothelial carcinoma. Histopathology 2019, 74, 483–493. [Google Scholar] [CrossRef]
- Breyer, J.; Wirtz, R.M.; Otto, W.; Erben, P.; Kriegmair, M.C.; Stoehr, R.; Eckstein, M.; Eidt, S.; Denzinger, S.; Burger, M.; et al. In stage pT1 non-muscle-invasive bladder cancer (NMIBC), high KRT20 and low KRT5 mRNA expression identify the luminal subtype and predict recurrence and survival. Virchows Arch. 2017, 470, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Eckstein, M.; Wirtz, R.M.; Gross-Weege, M.; Breyer, J.; Otto, W.; Stoehr, R.; Sikic, D.; Keck, B.; Eidt, S.; Burger, M.; et al. mRNA-expression of KRT5 and KRT20 defines distinct prognostic subgroups of muscle-invasive urothelial bladder cancer correlating with histological variants. Int. J. Mol. Sci. 2018, 19, 3396. [Google Scholar] [CrossRef] [PubMed]
- Biton, A.; Bernard-Pierrot, I.; Lou, Y.; Krucker, C.; Chapeaublanc, E.; Rubio-Perez, C.; Lopez-Bigas, N.; Kamoun, A.; Neuzillet, Y.; Gestraud, P.; et al. Independent component analysis uncovers the landscape of the bladder tumor transcriptome and reveals insights into luminal and basal subtypes. Cell Rep. 2014, 9, 1235–1245. [Google Scholar] [CrossRef]
- Tan, T.Z.; Rouanne, M.; Tan, K.T.; Huang, R.Y.; Thiery, J.P. Molecular subtypes of urothelial bladder cancer: Results from a meta-cohort analysis of 2411 tumors. Eur. Urol. 2018. [Google Scholar] [CrossRef]
- Rebouissou, S.; Bernard-Pierrot, I.; de Reynies, A.; Lepage, M.L.; Krucker, C.; Chapeaublanc, E.; Herault, A.; Kamoun, A.; Caillault, A.; Letouze, E.; et al. EGFR as a potential therapeutic target for a subset of muscle-invasive bladder cancers presenting a basal-like phenotype. Sci. Transl. Med. 2014, 6, 244ra91. [Google Scholar] [CrossRef] [PubMed]
- Dyrskjot, L.; Reinert, T.; Novoradovsky, A.; Zuiverloon, T.C.; Beukers, W.; Zwarthoff, E.; Malats, N.; Real, F.X.; Segersten, U.; Malmstrom, P.U.; et al. Analysis of molecular intra-patient variation and delineation of a prognostic 12-gene signature in non-muscle invasive bladder cancer; technology transfer from microarrays to PCR. Br. J. Cancer 2012, 107, 1392–1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelsey, K.T.; Hirao, T.; Schned, A.; Hirao, S.; Devi-Ashok, T.; Nelson, H.H.; Andrew, A.; Karagas, M.R. A population-based study of immunohistochemical detection of p53 alteration in bladder cancer. Br. J. Cancer 2004, 90, 1572–1576. [Google Scholar] [CrossRef] [Green Version]
- Bertz, S.; Otto, W.; Denzinger, S.; Wieland, W.F.; Burger, M.; Stohr, R.; Link, S.; Hofstadter, F.; Hartmann, A. Combination of CK20 and Ki-67 immunostaining analysis predicts recurrence, progression, and cancer-specific survival in pT1 urothelial bladder cancer. Eur. Urol. 2014, 65, 218–226. [Google Scholar] [CrossRef]
- Sjodahl, G.; Eriksson, P.; Liedberg, F.; Hoglund, M. Molecular classification of urothelial carcinoma: Global mRNA classification versus tumour-cell phenotype classification. J. Pathol. 2017, 242, 113–125. [Google Scholar] [CrossRef]
- Schaafsma, H.E.; Ramaekers, F.C.; van Muijen, G.N.; Lane, E.B.; Leigh, I.M.; Robben, H.; Huijsmans, A.; Ooms, E.C.; Ruiter, D.J. Distribution of cytokeratin polypeptides in human transitional cell carcinomas, with special emphasis on changing expression patterns during tumor progression. Am. J. Pathol. 1990, 136, 329–343. [Google Scholar]
- Volkmer, J.P.; Sahoo, D.; Chin, R.K.; Ho, P.L.; Tang, C.; Kurtova, A.V.; Willingham, S.B.; Pazhanisamy, S.K.; Contreras-Trujillo, H.; Storm, T.A.; et al. Three differentiation states risk-stratify bladder cancer into distinct subtypes. Proc. Natl. Acad. Sci. USA 2012, 109, 2078–2083. [Google Scholar] [CrossRef] [Green Version]
- Ho, P.L.; Kurtova, A.; Chan, K.S. Normal and neoplastic urothelial stem cells: Getting to the root of the problem. Nat. Rev. Urol. 2012, 9, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Sjodahl, G.; Lovgren, K.; Lauss, M.; Patschan, O.; Gudjonsson, S.; Chebil, G.; Aine, M.; Eriksson, P.; Mansson, W.; Lindgren, D.; et al. Toward a molecular pathologic classification of urothelial carcinoma. Am. J. Pathol. 2013, 183, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Patschan, O.; Sjodahl, G.; Chebil, G.; Lovgren, K.; Lauss, M.; Gudjonsson, S.; Kollberg, P.; Eriksson, P.; Aine, M.; Mansson, W.; et al. A molecular pathologic framework for risk stratification of stage T1 urothelial carcinoma. Eur. Urol. 2015, 68, 824–832. [Google Scholar] [CrossRef] [PubMed]
- Bryan, R.T. Cell adhesion and urothelial bladder cancer: The role of cadherin switching and related phenomena. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20140042. [Google Scholar] [CrossRef] [PubMed]
- Ramena, G.; Yin, Y.; Yu, Y.; Walia, V.; Elble, R.C. CLCA2 interactor EVA1 is required for mammary epithelial cell differentiation. PLoS ONE 2016, 11, e0147489. [Google Scholar] [CrossRef]
- Koivunen, J.; Aaltonen, V.; Koskela, S.; Lehenkari, P.; Laato, M.; Peltonen, J. Protein kinase C alpha/beta inhibitor Go6976 promotes formation of cell junctions and inhibits invasion of urinary bladder carcinoma cells. Cancer Res. 2004, 64, 5693–5701. [Google Scholar] [CrossRef]
- Ishida-Yamamoto, A.; Igawa, S.; Kishibe, M.; Honma, M. Clinical and molecular implications of structural changes to desmosomes and corneodesmosomes. J. Dermatol. 2018, 45, 385–389. [Google Scholar] [CrossRef]
- Jacquemet, G.; Hamidi, H.; Ivaska, J. Filopodia in cell adhesion, 3D migration and cancer cell invasion. Curr. Opin. Cell Biol. 2015, 36, 23–31. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Z.; Luan, X.; Zha, J.; Li, Z.; Wu, L.; Yan, Y.; Wang, H.; Hou, D.; Huang, L.; Huang, F.; et al. TNF-alpha inhibits the migration of oral squamous cancer cells mediated by miR-765-EMP3-p66Shc axis. Cell Signal 2017, 34, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Brunner, A.; Tzankov, A. The role of structural extracellular matrix proteins in urothelial bladder cancer (review). Biomark. Insights 2007, 2, 418–427. [Google Scholar] [CrossRef] [PubMed]
- Kramer, M.W.; Waalkes, S.; Serth, J.; Hennenlotter, J.; Tezval, H.; Stenzl, A.; Kuczyk, M.A.; Merseburger, A.S. Decreased galectin-8 is a strong marker for recurrence in urothelial carcinoma of the bladder. Urol. Int. 2011, 87, 143–150. [Google Scholar] [CrossRef]
- Szekely, E.; Torzsok, P.; Riesz, P.; Korompay, A.; Fintha, A.; Szekely, T.; Lotz, G.; Nyirady, P.; Romics, I.; Timar, J.; et al. Expression of claudins and their prognostic significance in noninvasive urothelial neoplasms of the human urinary bladder. J. Histochem. Cytochem. 2011, 59, 932–941. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y. Wnt/Planar cell polarity signaling: A new paradigm for cancer therapy. Mol. Cancer Ther. 2009, 8, 2103–2109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, C.; Zou, H.; Zhang, J.; Wang, J.; Liu, H. An integrated methylation and gene expression microarray analysis reveals significant prognostic biomarkers in oral squamous cell carcinoma. Oncol. Rep. 2018, 40, 2637–2647. [Google Scholar] [CrossRef]
- Sun, H.T.; Cheng, S.X.; Tu, Y.; Li, X.H.; Zhang, S. FoxQ1 promotes glioma cells proliferation and migration by regulating NRXN3 expression. PLoS ONE 2013, 8, e55693. [Google Scholar] [CrossRef] [PubMed]
- O’Sullivan, A.G.; Mulvaney, E.P.; Kinsella, B.T. Regulation of protein kinase C-related kinase (PRK) signalling by the TPalpha and TPbeta isoforms of the human thromboxane A2 receptor: Implications for thromboxane- and androgen- dependent neoplastic and epigenetic responses in prostate cancer. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 838–856. [Google Scholar] [CrossRef]
- Nagaishi, M.; Kim, Y.H.; Mittelbronn, M.; Giangaspero, F.; Paulus, W.; Brokinkel, B.; Vital, A.; Tanaka, Y.; Nakazato, Y.; Legras-Lachuer, C.; et al. Amplification of the STOML3, FREM2, and LHFP genes is associated with mesenchymal differentiation in gliosarcoma. Am. J. Pathol. 2012, 180, 1816–1823. [Google Scholar] [CrossRef]
- Fang, D.; Kitamura, H. Cancer stem cells and epithelial-mesenchymal transition in urothelial carcinoma: Possible pathways and potential therapeutic approaches. Int. J. Urol. 2018, 25, 7–17. [Google Scholar] [CrossRef]
- Sun, Y.; Jiang, F.; Pan, Y.; Chen, X.; Chen, J.; Wang, Y.; Zheng, X.; Zhang, J. XBP1 promotes tumor invasion and is associated with poor prognosis in oral squamous cell carcinoma. Oncol. Rep. 2018, 40, 988–998. [Google Scholar] [CrossRef]
- Takahashi, H.; Nakatsuji, H.; Takahashi, M.; Avirmed, S.; Fukawa, T.; Takemura, M.; Fukumori, T.; Kanayama, H. Up-regulation of plakophilin-2 and Down-regulation of plakophilin-3 are correlated with invasiveness in bladder cancer. Urology 2012, 79, 240.e1–e8. [Google Scholar] [CrossRef]
- Miyake, H.; Hara, I.; Yamanaka, K.; Gohji, K.; Arakawa, S.; Kamidono, S. Increased angiogenin expression in the tumor tissue and serum of urothelial carcinoma patients is related to disease progression and recurrence. Cancer 1999, 86, 316–324. [Google Scholar] [CrossRef] [Green Version]
- Frantzi, M.; Klimou, Z.; Makridakis, M.; Zoidakis, J.; Latosinska, A.; Borras, D.M.; Janssen, B.; Giannopoulou, I.; Lygirou, V.; Lazaris, A.C.; et al. Silencing of Profilin-1 suppresses cell adhesion and tumor growth via predicted alterations in integrin and Ca2+ signaling in T24M-based bladder cancer models. Oncotarget 2016, 7, 70750–70768. [Google Scholar] [CrossRef] [Green Version]
- Kowalski, M.; Entwistle, J.; Cizeau, J.; Niforos, D.; Loewen, S.; Chapman, W.; MacDonald, G.C. A Phase I study of an intravesically administered immunotoxin targeting EpCAM for the treatment of nonmuscle-invasive bladder cancer in BCGrefractory and BCG-intolerant patients. Drug Des. Dev. Ther. 2010, 4, 313–320. [Google Scholar] [CrossRef]
- Desai, S.; Lim, S.D.; Jimenez, R.E.; Chun, T.; Keane, T.E.; McKenney, J.K.; Zavala-Pompa, A.; Cohen, C.; Young, R.H.; Amin, M.B. Relationship of cytokeratin 20 and CD44 protein expression with WHO/ISUP grade in pTa and pT1 papillary urothelial neoplasia. Mod. Pathol. 2000, 13, 1315–1323. [Google Scholar] [CrossRef]
- Delire, B.; Starkel, P. The Ras/MAPK pathway and hepatocarcinoma: Pathogenesis and therapeutic implications. Eur. J. Clin. Investig. 2015, 45, 609–623. [Google Scholar] [CrossRef]
- Kim, J.H.; Park, S.; Chung, H.; Oh, S. Wnt5a attenuates the pathogenic effects of the Wnt/beta-catenin pathway in human retinal pigment epithelial cells via down-regulating beta-catenin and Snail. BMB Rep. 2015, 48, 525–530. [Google Scholar] [CrossRef]
- Wu, X.R. Urothelial tumorigenesis: A tale of divergent pathways. Nat. Rev. Cancer 2005, 5, 713–725. [Google Scholar] [CrossRef]
- Sethi, G.; Sung, B.; Aggarwal, B.B. TNF: A master switch for inflammation to cancer. Front. Biosci. 2008, 13, 5094–5107. [Google Scholar] [CrossRef]
- Van Batavia, J.; Yamany, T.; Molotkov, A.; Dan, H.; Mansukhani, M.; Batourina, E.; Schneider, K.; Oyon, D.; Dunlop, M.; Wu, X.R.; et al. Bladder cancers arise from distinct urothelial sub-populations. Nat. Cell Biol. 2014, 16, 982–991. [Google Scholar] [CrossRef]
- Martin, J.A.; Wang, Z. Next-generation transcriptome assembly. Nat. Rev. Genet. 2011, 12, 671–682. [Google Scholar] [CrossRef] [Green Version]
- Babraham Bioinformatics. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 18 December 2018).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef]









| Variables | Group 1 | Group 2 | Group 3 | p-value |
|---|---|---|---|---|
| Number (%) | 5 (100) | 5 (100) | 5 (100) | |
| Age | 1.000 | |||
| ≥69 | 3 (60) | 3 (60) | 3 (60) | |
| <69 | 2 (40) | 2 (40) | 2 (40) | |
| Sex | 1.000 | |||
| Male | 3 (60) | 4 (80) | 3 (60) | |
| Female | 2 (40) | 1 (20) | 2 (40) | |
| Size | 0.725 | |||
| ≥3.6 | 2 (40) | 1 (20) | 0 (0) | |
| <3.6 | 3 (60) | 4 (80) | 5 (100) | |
| Organ | 0.301 | |||
| Pelvis/calyx | 4 (80) | 1 (20) | 3 (60) | |
| Ureter | 1 (20) | 4 (80) | 2 (40) | |
| Stage | 0.800 | |||
| T1 | 4 (80) | 2 (40) | 3 (60) | |
| Ta | 1 (20) | 3 (60) | 2 (40) | |
| CIS | 1.000 | |||
| Present | 2 (40) | 1 (20) | 2 (40) | |
| Absent | 3 (60) | 4 (80) | 3 (60) | |
| IHC score (mean ± SD) 1 | ||||
| CK5/6 | 6.4 ± 2.41 | 7.4 ± 2.41 | 1 ± 0.00 | 0.001 |
| CK20 | 1.0 ± 0.00 | 8.4 ± 1.52 | 10 ± 0.00 | <0.001 |
| CD44 | 6.0 ± 2.83 | 5.4 ± 3.21 | 3.0 ± 2.55 | 0.162 |
| p53 | 7.5 ± 3.70 | 6.3 ± 2.22 | 4.8 ± 3.55 | 0.428 |
| CK14 | 1.4 ± 0.89 | 1.0 ± 0.00 | 1.0 ± 0.00 | 1.000 |
| GATA3 | 8.0 ± 3.94 | 10.0 ± 0.00 | 10.0 ± 0.00 | 0.286 |
| FOXA1 | 6.4 ± 4.51 | 10.0 ± 0.00 | 8.4 ± 1.95 | 0.066 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, M.; Lee, J.H.; Kim, B.; Park, J.H.; Moon, K.C. Transcriptional Analysis of Immunohistochemically Defined Subgroups of Non-Muscle-Invasive Papillary High-Grade Upper Tract Urothelial Carcinoma. Int. J. Mol. Sci. 2019, 20, 570. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20030570
Jung M, Lee JH, Kim B, Park JH, Moon KC. Transcriptional Analysis of Immunohistochemically Defined Subgroups of Non-Muscle-Invasive Papillary High-Grade Upper Tract Urothelial Carcinoma. International Journal of Molecular Sciences. 2019; 20(3):570. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20030570
Chicago/Turabian StyleJung, Minsun, Jeong Hoon Lee, Bohyun Kim, Jeong Hwan Park, and Kyung Chul Moon. 2019. "Transcriptional Analysis of Immunohistochemically Defined Subgroups of Non-Muscle-Invasive Papillary High-Grade Upper Tract Urothelial Carcinoma" International Journal of Molecular Sciences 20, no. 3: 570. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20030570