Analysis of the miRNA Expression Profiles in the Zearalenone-Exposed TM3 Leydig Cell Line
Abstract
:1. Introduction
2. Results
2.1. Effect of ZEN on the Proliferation of TM3 Leydig Cells
2.2. Differential Expression of miRNAs in the ZEN Exposure Groups
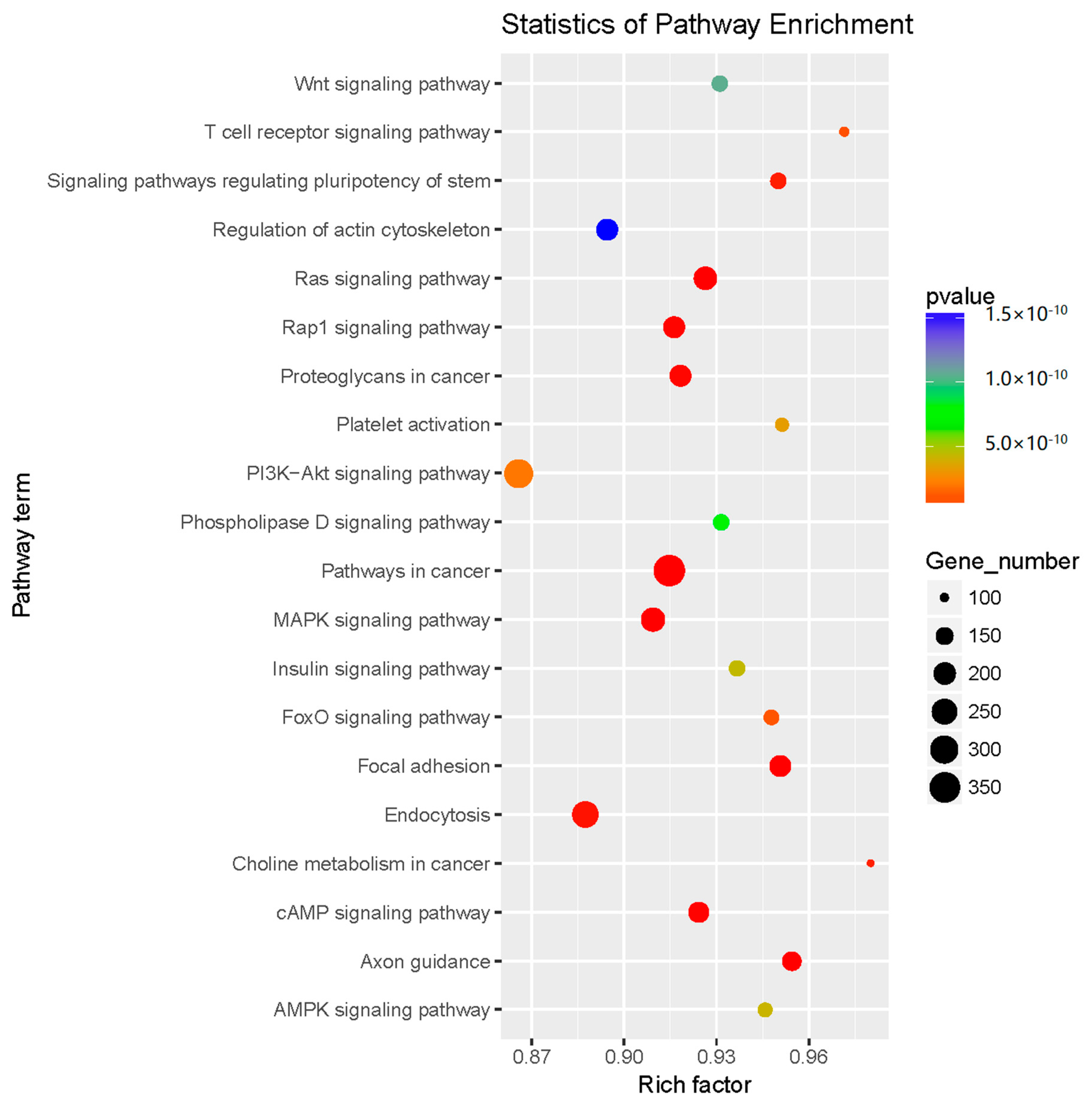
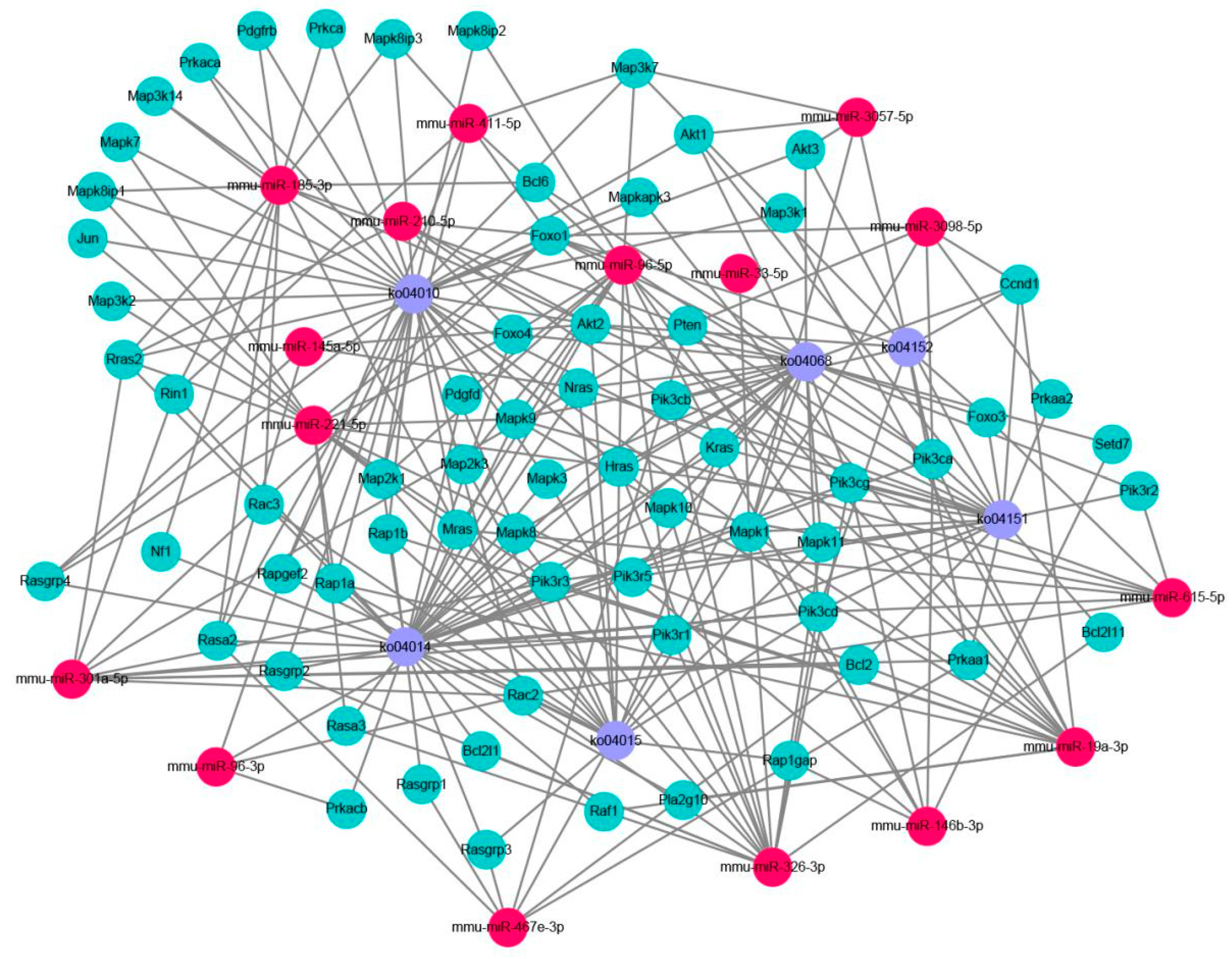
2.3. Prediction and Functional Classification of Target Genes
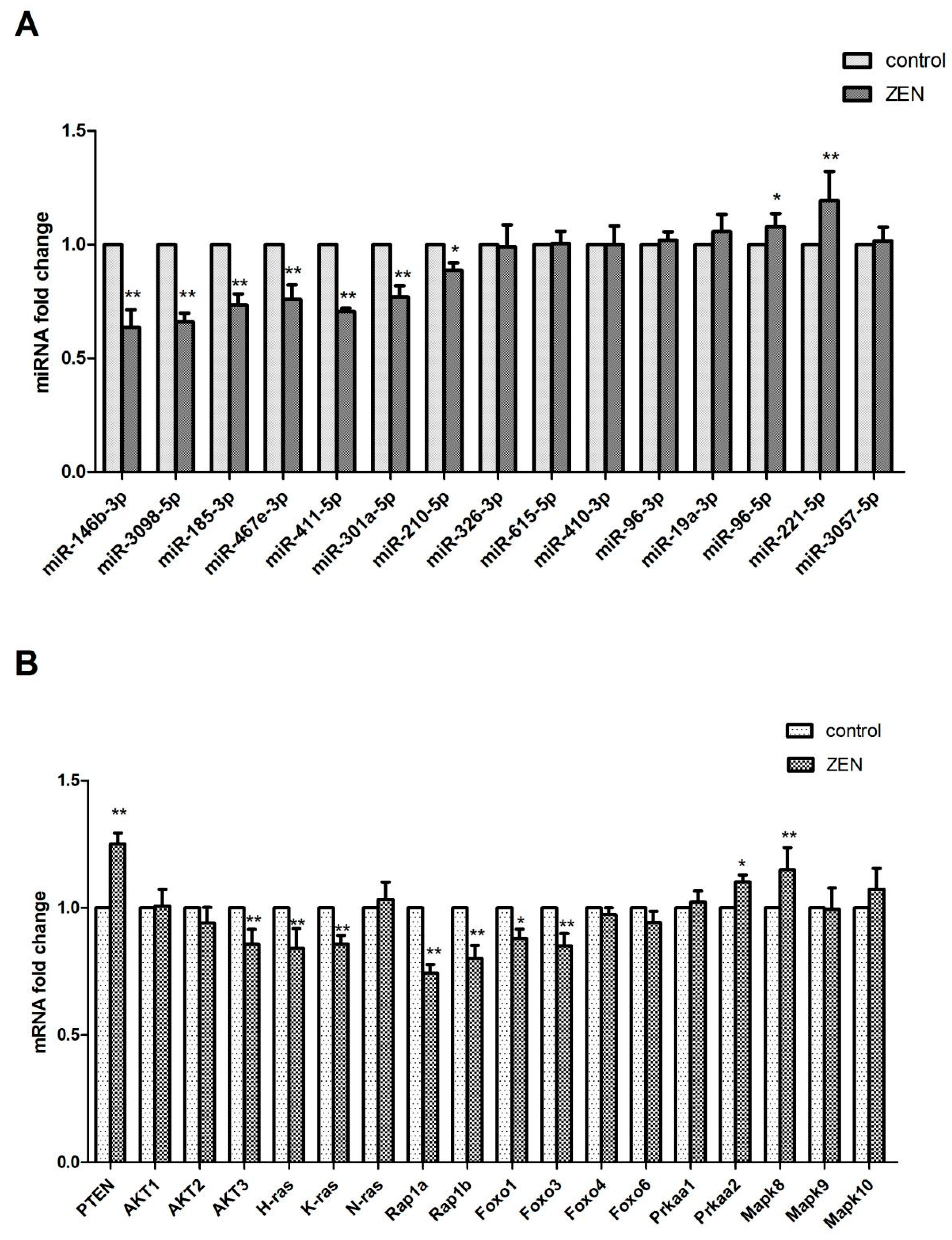
2.4. Validation of the Differentially Expressed miRNAs
2.5. Verification of the miRNA Target Genes
3. Discussion
4. Materials and Method
4.1. Cell Culture and ZEN Exposure
4.2. Cell Viability Assay
4.3. Small RNA Sequencing and Bioinformatics Analysis
4.4. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Scudamore, K.A.; Patel, S. Survey for aflatoxins, ochratoxin a, zearalenone and fumonisins in maize imported into the United Kingdom. Food Addit. Contam. 2000, 17, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Zinedine, A.; Soriano, J.M.; Moltó, J.C.; Mañes, J. Review on the toxicity, occurrence, metabolism, detoxification, regulations and intake of zearalenone: Ans oestrogenic mycotoxin. Food Chem. Toxicol. 2007, 45, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Olsen, M.; Pettersson, H.; Kiessling, K.H. Reduction of zearalenone to zearalenol in female rat liver by 3 alpha-hydroxysteroid dehydrogenase. Basic Clin. Pharmacol. Toxicol. 2010, 48, 157–161. [Google Scholar]
- Turcotte, J.C.; Hunt, P.J.B.; Blaustein, J.D. Estrogenic effects of zearalenone on the expression of progestin receptors and sexual behavior in female rats. Horm. Behav. 2005, 47, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Benzoni, E.; Minervini, F.; Giannoccaro, A.; Fornelli, F.; Vigo, D.; Visconti, A. Influence of in vitro exposure to mycotoxin zearalenone and its derivatives on swine sperm quality. Reprod. Toxicol. 2008, 25, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Wang, G.X.; Liu, J.L.; Fan, J.J.; Cui, S. Toxic effects of zearalenone and its derivatives alpha-zearalenol on male reproductive system in mice. Reprod. Toxicol. 2007, 24, 381–387. [Google Scholar] [CrossRef] [PubMed]
- Yuanyuan, Z.; Zhiqiang, J.; Shutong, Y.; Anshan, S.; Rui, G.; Zhe, Q.; Min, L.; Shaoping, N. Toxic effects of maternal zearalenone exposure on uterine capacity and fetal development in gestation rats. Reprod. Sci. 2014, 21, 743–753. [Google Scholar]
- Shi, B.; Su, Y.; Chang, S.; Sun, Y.; Meng, X.; Shan, A. Vitamin c protects piglet liver against zearalenone-induced oxidative stress by modulating expression of nuclear receptors pxr and car and their target genes. Food Funct. 2017, 8, 3675–3687. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Lv, Y.; Ren, S.; Shao, M.; Shen, T.; Huang, K.; Zhou, J.; Yan, L.; Song, S. Zearalenone (zea)-induced intestinal inflammation is mediated by the nlrp3 inflammasome. Chemosphere 2017, 190, 272. [Google Scholar] [CrossRef] [PubMed]
- Ben, S.I.; Boussabbeh, M.; Da, S.J.; Guilbert, A.; Bacha, H.; Abidessefi, S.; Lemaire, C. Sirt1 protects cardiac cells against apoptosis induced by zearalenone or its metabolites α- and β-zearalenol through an autophagy-dependent pathway. Toxicol. Appl. Pharmacol. 2016, 314, 82–90. [Google Scholar]
- Long, M.; Yang, S.H.; Shi, W.; Li, P.; Guo, Y.; Guo, J.; He, J.B.; Zhang, Y. Protective effect of proanthocyanidin on mice sertoli cell apoptosis induced by zearalenone via the nrf2/are signalling pathway. Environ. Sci. Pollut. Res. Int. 2017, 24, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Donadeu, F.X.; Sontakke, S.D.; Ioannidis, J. Microrna indicators of follicular steroidogenesis. Reprod. Fertil. Dev. 2016, 29, 906–912. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.M.; Chaturvedi, G.; Tong, S.K.; Nusrin, S.; Giesy, J.P.; Wu, R.S.; Kong, R.Y. Evidence for microrna-mediated regulation of steroidogenesis by hypoxia. Environ. Sci. Technol. 2015, 49, 1138–1147. [Google Scholar] [CrossRef] [PubMed]
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Switching from repression to activation: Micrornas can up-regulate translation. Science 2007, 318, 1931–1934. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Kim, B.; Kim, V.N. Re-evaluation of the roles of drosha, exportin 5, and dicer in microrna biogenesis. Proc. Natl. Acad. Sci. USA 2016, 113, E1881. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Gao, S.; Chen, S.; Chen, L.; Zhao, Y.; Jiang, Y.; Zheng, X.; Zhou, X. Differential expression of micrornas in luteinising hormone-treated mouse tm3 leydig cells. Andrologia 2017, 50, e12824. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Li, C.; Xu, Y.; Chen, S.; Zhao, Y.; Chen, L.; Jiang, Y.; Liu, Z.; Fan, R.; Sun, L. Differential expression of micrornas in tm3 leydig cells of mice treated with brain-derived neurotrophic factor. Cell Biochem. Funct. 2017, 35, 364–371. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Pan, S.; Wang, G.; Wang, Y.J.; Liu, Q.; Gu, J.; Yuan, Y.; Liu, X.Z.; Liu, Z.P.; Bian, J.C. Zearalenone impairs the male reproductive system functions via inducing structural and functional alterations of sertoli cells. Environ. Toxicol. Pharmacol. 2016, 42, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Chin, L.; Tam, A.; Pomerantz, J.; Wong, M.; Holash, J.; Bardeesy, N.; Shen, Q.; O’Hagan, R.; Pantginis, J.; Zhou, H. Essential role for oncogenic ras in tumour maintenance. Nature 1999, 400, 468–472. [Google Scholar] [CrossRef] [PubMed]
- Stephen, A.; Esposito, D.; Bagni, R.; Mccormick, F. Dragging ras back in the ring. Cancer Cell 2014, 25, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.; Brock, E.J.; Ji, K.; Mattingly, R.R. Ras and rap1: A tale of two gtpases. In Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2018. [Google Scholar]
- Hampl, R.; Kubátová, J.; Stárka, L. Steroids and endocrine disruptors-history, recent state of art and open questions. J. Steroid Biochem. Mol. Biol. 2016, 155, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Huang, Q.; Pan, S.; Fan, W.; Wang, G.; Yuan, Y.; Gu, J.; Liu, X.; Liu, Z.; Bian, J. Regulation of oncogenes and gap junction intercellular communication during the proliferative response of zearalenone in tm3 cells. Hum. Exp. Toxicol. 2016, 36. [Google Scholar] [CrossRef] [PubMed]
- Miyata, S.; Fukuda, Y.; Tojima, H.; Matsuzaki, K.; Kitanaka, S.; Sawada, H. Mechanism of the inhibition of leukemia cell growth and induction of apoptosis through the activation of atr and pten by the topoisomerase inhibitor 3ez, 20ac-ingenol. Leuk. Res. 2015, 39, 927–932. [Google Scholar] [CrossRef] [PubMed]
- Long, Z.W.; Wu, J.H.; Hong, C.; Wang, Y.N.; Zhou, Y. Mir-374b promotes proliferation and inhibits apoptosis of human gist cells by inhibiting pten through activation of the pi3k/akt pathway. Mol. Cells 2018, 41, 532–544. [Google Scholar] [PubMed]
- Jiang, Y.; Chang, H.; Chen, G. Effects of microrna-20a on the proliferation, migration and apoptosis of multiple myeloma via the pten/pi3k/akt signaling pathway. Oncol. Lett. 2018. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Song, Y.; Li, G.; Ren, J.; Xie, J.; Zhang, Y.; Gao, F.; Mu, J.; Dai, J. Downregulation of microrna-21 expression inhibits proliferation, and induces g1 arrest and apoptosis via the pten/akt pathway in skm-1 cells. Mol. Med. Re. 2018, 18, 2771–2779. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhang, L.; Zhou, Z.Q.; Ren, Y.Q.; Sun, L.N.; Man, Y.L.; Ma, Z.W.; Wang, Z.K. Effects of long non-coding rna linc00963 on renal interstitial fibrosis and oxidative stress of rats with chronic renal failure via the foxo signaling pathway. Cell. Physiol. Biochem. 2018, 46, 815–828. [Google Scholar] [CrossRef]
- Guo, F.; Yu, X.; Xu, A.; Xu, J.; Wang, Q.; Guo, Y.; Wu, X.; Tang, Y.; Ding, Z.; Zhang, Y. Japanese encephalitis virus induces apoptosis by inhibiting foxo signaling pathway. Vet. Microbiol. 2018, 220, 73. [Google Scholar] [CrossRef]
- Long, M.; Yang, S.H.; Han, J.X.; Li, P.; Zhang, Y.; Dong, S.; Chen, X.; Guo, J.; Wang, J.; He, J.B. The protective effect of grape-seed proanthocyanidin extract on oxidative damage induced by zearalenone in kunming mice liver. Int. J. Mol. Sci. 2016, 17, 808. [Google Scholar] [CrossRef]
- Zheng, W.L.; Wang, B.J.; Wang, L.; Shan, Y.P.; Zou, H.; Song, R.L.; Wang, T.; Gu, J.H.; Yuan, Y.; Liu, X.Z. Ros-mediated cell cycle arrest and apoptosis induced by zearalenone in mouse sertoli cells via er stress and the atp/ampk pathway. Toxins 2018, 10, 24. [Google Scholar] [CrossRef]
- Pistol, G.C.; Braicu, C.; Motiu, M.; Gras, M.A.; Marin, D.E.; Stancu, M.; Calin, L.; Israel-Roming, F.; Berindan-Neagoe, I.; Taranu, I. Zearalenone mycotoxin affects immune mediators, mapk signalling molecules, nuclear receptors and genome-wide gene expression in pig spleen. PLoS ONE 2015, 10, e0127503. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, N.; Tong, J.; Pan, J.; Long, M.; Li, P. Transcriptome analysis to identify the ras and rap1 signal pathway genes involved in the response of tm3 leydig cells exposed to zearalenone. Environ. Sci. Pollut. Res. Int. 2018, 25, 31230–31239. [Google Scholar] [CrossRef] [PubMed]


| miRNA Name | log2 (Fold Change) a | p Value (Chi_Square_2×2) | p Value (Fisher Test) |
|---|---|---|---|
| miR-146b-3p | −2.5 | 3.01 × 10−3 | 9.29 × 10−3 |
| miR-3098-5p | −2.26 | 8.57 × 10−3 | 3.38 × 10−2 |
| miR-185-3p | −2.12 | 5.42 × 10−4 | 1.11 × 10−3 |
| miR-467e-3p | −2.09 | 3.24 × 10−3 | 2.43 × 10−3 |
| miR-411-5p | −1.95 | 6.82 × 10−18 | 1.09 × 10−17 |
| miR-301a-5p | −1.71 | 2.14 × 10−6 | 3.17 × 10−6 |
| miR-210-5p | −1.61 | 1.23 × 10−10 | 3.57 × 10−10 |
| miR-326-3p | −1.30 | 2.59 × 10−4 | 6.30 × 10−4 |
| miR-615-5p | −1.23 | 9.62 × 10−51 | 2.25 × 10−49 |
| miR-410-3p | −1.17 | 8.96 × 10−4 | 1.79 × 10−3 |
| miR-96-3p | −1.06 | 5.13 × 10−3 | 1.04 × 10−2 |
| miR-19a-3p | 1.21 | 1.02 × 10−7 | 6.96 × 10−8 |
| miR-96-5p | 1.47 | 3.67 × 10−47 | 1.28 × 10−49 |
| miR-221-5p | 2.0 | 1.63 × 10−62 | 2.13 × 10−68 |
| miR-3057-5p | 2.01 | 4.49 × 10−11 | 8.80 × 10−12 |
| Gene | Primer Sequence (5′-3′) | Accession No |
|---|---|---|
| β-actin | Forward: CTGTCCCTGTATGCCTCTG Reverse: TTGATGTCACGCACGATT | BC_138614.1 |
| N-ras | Forward: GGTTGGAGCAGGTGGTGTT Reverse: TTTCGGTAAGAATCCTCTATG | NM_010937.2 |
| K-ras | Forward: TGCCTTCTAGAACAGTAGACAC Reverse: CTTTGCTGAGGTCTCAATGAAC | NM_021284.6 |
| H-ras | Forward: GCATCCCCTACATTGAAACATC Reverse: CAATTTATGCTGCCGAATCTCA | NM_001130443.1 |
| Pten | Forward: TGGATTCGACTTAGACTTGACC Reverse: TCACTTAGCCATTGGTCAAGAT | NM_008960.2 |
| Akt1 | Forward: TGCACAAACGAGGGGAATATAT Reverse: CGTTCCTTGTAGCCAATAAAGG | NM_001165894.1 |
| Akt2 | Forward: TCGATTATCTCAAACTCCTCGG Reverse: CGACTTCATCCTTTGCAATGAT | NM_001110208.2 |
| Akt3 | Forward: GGGGTGGAACAGTAAAGACA Reverse: GCATTATGAGCAGTGGAGG | NM_011785.4 |
| Rap1a | Forward: ATTCCTACAGAAAGCAAGTCGA Reverse: ATCTTCTGTGTCTTTAACCCGT | NM_145541.5 |
| Rap1b | Forward: AAGCAAGTTGAAGTAGATGCAC Reverse: CATCATCAGTGTCTTTAACCCG | NM_024457.2 |
| Foxo1 | Forward: GATCTACGAGTGGATGGTGAAG Reverse: GACAGATTGTGGCGAATTGAAT | NM_019739.3 |
| Foxo3 | Forward: TCACTGTATTCAGCTAGTGCAA Reverse: ATGATGGACTCCATGTCACATT | NM_019740.2 |
| Foxo4 | Forward: GAATCCTGGGGGCTGTAAC Reverse: GCTGATGAGTTCTGCATATGAC | NM_018789.2 |
| Foxo6 | Forward: GAAAGCGAAGAGCTCCCGAC Reverse: GTGCCGAATGGAGTTCTTCCAG | NM_194060.1 |
| Prkaa1 | Forward: GGACTTACTTGTTGGATTTCCG Reverse: CCTTTGGCAAGATCGATAGTTG | NM_001013367.3 |
| Prkaa2 | Forward: GTGGTGACCCTCAAGACCAG Reverse: GTGGTTTCAAGCCTGGAGGA | NM_001356568.1 |
| Mapk8 | Forward: TTGAAAACAGGCCTAAATACGC Reverse: GTTTGTTATGCTCTGAGTCAGC | NM_001310452.1 |
| Mapk9 | Forward: GTGGAAAACAGACCAAAGTACC Reverse: CATGCTCTCTTTCTTCCAACTG | NM_001163671.1 |
| Mapk10 | Forward: CACGAGCGGATGTCTTACT Reverse: TTGACTACAATGTTACTGGGTT | NM_001081567.2 |
| miRNA | Primer sequence (5′-3′) |
|---|---|
| U6 | RT: CGCTTCACGAATTTGCGTGTCAT Forward: GCTTCGGCAGCACATATACTAAAAT Reverse: CGCTTCACGAATTTGCGTGTCAT |
| miR-146b-3p | RT: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACCAGA Forward: GCGGCCCTAGGGACTCAGT Reverse: AGTGCAGGGTCCGAGGTATT |
| miR-185-3p | RT: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACCAGA Forward: CGAGGGGCTGGCTTTCC Reverse: AGTGCAGGGTCCGAGGTATT |
| miR-3098-5p | RT: TGACCGTCTGTATGGTTGTTCACGACTCCTTCACCCTATCCAACCATACAGACGGTCAGCTCCTAC Forward: GGGTCCTAACAGCAGGAGTA Reverse: TATGGTTGTTCACGACTCCTTCAC |
| miR-467e-3p | RT: TGACCGTCTGTATGGTTGTTCACGACTCCTTCACCCTATCCAACCATACAGACGGTCAATATAGGTG Forward: GGGATATACATACACACAC Reverse: TATGGTTGTTCACGACTCCTTCAC |
| miR-411-5p | RT: TGACCGTCTGTATGGTTGTTCACGACTCCTTCACCCTATCCAACCATACAGACGGTCACGTACGCT Forward: GGGTAGTAGACCGTATAGC Reverse: TATGGTTGTTCACGACTCCTTCAC |
| miR-326-3p | RT: TGACCGTCTGTATGGTTGTTCACGACTCCTTCACCCTATCCAACCATACAGACGGTCAACTGGAGG Forward: GGGCCTCTGGGCCCTTCCT Reverse: TATGGTTGTTCACGACTCCTTCAC |
| miR-615-5p | RT: TGACCGTCTGTATGGTTGTTCACGACTCCTTCACCCTATCCAACCATACAGACGGTCAGATCCGAG Forward: GGGGGTCCCCGGTGCT Reverse: TATGGTTGTTCACGACTCCTTCAC |
| miR-410-3p | RT: TGACCGTCTGTATGGTTGTTCACGACTCCTTCACCCTATCCAACCATACAGACGGTCAACAGGCCA Forward: GGGAATATAACACAGATGG Reverse: TATGGTTGTTCACGACTCCTTCAC |
| miR-96-3p | RT: TGACCGTCTGTATGGTTGTTCACGACTCCTTCACCCTATCCAACCATACAGACGGTCAATATTGGC Forward: GGGCAATCATGTGTAGTGC Reverse: TATGGTTGTTCACGACTCCTTCAC |
| miR-19a-3p | RT: TGACCGTCTGTATGGTTGTTCACGACTCCTTCACCCTATCCAACCATACAGACGGTCATCAGTTTTG Forward: GGGTGTGCAAATCTATGCAA Reverse: TATGGTTGTTCACGACTCCTTCAC |
| miR-3057-5p | RT: TGACCGTCTGTATGGTTGTTCACGACTCCTTCACCCTATCCAACCATACAGACGGTCAATCCCGCA Forward: GGGATTGGAGCTGAGATTCTG Reverse: TATGGTTGTTCACGACTCCTTCAC |
| miR-301a-5p | RT: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGTAGT Forward: CGCGGCTCTGACTTTATTGC Reverse: AGTGCAGGGTCCGAGGTATT |
| miR-221-5p | RT: GAGGTATTCGCACTGGATACGACACAGAA Forward: GCGACCTGGCATACAATGTAGAT Reverse: AGTGCAGGGTCCGAGGTATT |
| miR-96-5p | RT: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGCAAA Forward: GCGTTTGGCACTAGCACATT Reverse: AGTGCAGGGTCCGAGGTATT |
| miR-210-5p | RT: TGACCGTCTGTATGGTTGTTCACGACTCCTTCACCCTATCCAACCATACAGACGGTCACAGTGTGC Forward: GGGAGCCACTGCCCACCGC Reverse: TATGGTTGTTCACGACTCCTTCAC |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Wu, W.; Li, L.; He, J.; Huang, S.; Chen, S.; Chen, J.; Long, M.; Yang, S.; Li, P. Analysis of the miRNA Expression Profiles in the Zearalenone-Exposed TM3 Leydig Cell Line. Int. J. Mol. Sci. 2019, 20, 635. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20030635
Wang M, Wu W, Li L, He J, Huang S, Chen S, Chen J, Long M, Yang S, Li P. Analysis of the miRNA Expression Profiles in the Zearalenone-Exposed TM3 Leydig Cell Line. International Journal of Molecular Sciences. 2019; 20(3):635. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20030635
Chicago/Turabian StyleWang, Mingyang, Weiwei Wu, Lin Li, Jianbin He, Sheng Huang, Si Chen, Jia Chen, Miao Long, Shuhua Yang, and Peng Li. 2019. "Analysis of the miRNA Expression Profiles in the Zearalenone-Exposed TM3 Leydig Cell Line" International Journal of Molecular Sciences 20, no. 3: 635. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20030635





