A Thermostable Monoacylglycerol Lipase from Marine Geobacillus sp. 12AMOR1: Biochemical Characterization and Mutagenesis Study
Abstract
:1. Introduction
2. Results and Discussion
2.1. Gene Sequence Identification and Recombinant Protein Production
2.2. Biochemical Characterization of Monoacylglycerol Lipase from Geobacillus sp. (GMGL)
2.2.1. Effect of Temperature on Lipase Activity and Thermostability
2.2.2. Effect of pH on Lipase Activity of GMGL
2.2.3. Effects of Metal Ions and Solvents on Lipase Activity of GMGL
2.2.4. Substrate Specificity of GMGL
2.3. Structure Analysis of GMGL
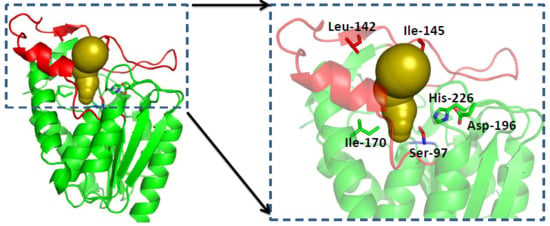
2.3.1. Structure Model of GMGL
2.3.2. Molecular Basis for the Acyl Chain Length Selectivity
3. Materials and Methods
3.1. Materials
3.2. Expression and Purification of GMGL
3.3. Biochemical Characterization of GMGL
3.3.1. Activity Determination
3.3.2. Effects of Temperature on the Lipase Activity of GMGL
3.3.3. Effect of pH on Lipase Activity of GMGL
3.3.4. Substrate Specificity of GMGL Lipase
3.3.5. Effect of Metal Ions, Detergents and Organic Solvents on the Lipase Activity of GMGL
3.3.6. Determination of Enzyme Kinetic Parameters
3.4. Hydrolysis Activity of Triacylglycerol (TAG) and 2,2-Dilauric Acid Glyceride (DAG)
3.5. Esterification Activity of GMGL
3.6. Homology Modeling and Docking Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| GMGL | Geobacillus sp. monoacylglycerol Lipase |
| MGL | Monoacylglycerol Lipase |
| MAG | Monoacylglycerol |
| DAG | Diacylglycerol |
| TAG | Triacylglycerol |
| WT | Wild type |
| IPTG | Isopropyl β-d-Thiogalactoside |
| pNP-C4 | 4-Nitrophenyl butyrate |
| pNP-C6 | 4-Nitrophenyl caproate |
| pNP-C8 | 4-Nitrophenyl octanoate |
| pNP-C10 | 4-Nitrophenyl decanoate |
| pNP-C12 | 4-Nitrophenyl laurate |
| pNP-C14 | 4-Nitrophenyl myristate |
| pNP-C16 | 4-Nitrophenyl palmitate |
| pNP-C18 | 4-Nitrophenyl stearate |
References
- Rudkowska, I.; Roynette, C.E.; Demonty, I.; Vanstone, C.A.; Jew, S.; Jones, P.J. Diacylglycerol: Efficacy and mechanism of action of an anti-obesity agent. Obes. Res. 2005, 13, 1864–1876. [Google Scholar] [CrossRef] [PubMed]
- Khaddaj-Mallat, R.; Morin, C.; Rousseau, E. Novel n-3 PUFA monoacylglycerides of pharmacological and medicinal interest: Anti-inflammatory and anti-proliferative effects. Eur. J. Pharmacol. 2016, 792, 70–77. [Google Scholar] [CrossRef] [PubMed]
- von der Haar, D.; Stabler, A.; Wichmann, R.; Schweiggert-Weisz, U. Enzyme-assisted process for DAG synthesis in edible oils. Food Chem. 2015, 176, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Cotes, K.; Dhouib, R.; Douchet, I.; Chahinian, H.; de Caro, A.; Carriere, F.; Canaan, S. Characterization of an exported monoglyceride lipase from Mycobacterium tuberculosis possibly involved in the metabolism of host cell membrane lipids. Biochem. J. 2007, 408, 417–427. [Google Scholar] [CrossRef] [PubMed]
- McPherson, J.C.; Askins, R.E.; Pope, J.L. The specificity of an intestinal lipase for monoglycerides. Exp. Biol. Med. 1962, 110, 744–748. [Google Scholar] [CrossRef]
- Aschauer, P.; Zimmermann, R.; Breinbauer, R.; Pavkov-Keller, T.; Oberer, M. The crystal structure of monoacylglycerol lipase from M. tuberculosis reveals the basis for specific inhibition. Sci. Rep. 2018, 8, 8948. [Google Scholar] [CrossRef] [PubMed]
- Aschauer, P.; Rengachari, S.; Lichtenegger, J.; Schittmayer, M.; Das, K.M.; Mayer, N.; Breinbauer, R.; Birner-Gruenberger, R.; Gruber, C.C.; Zimmermann, R.; et al. Crystal structure of the Saccharomyces cerevisiae monoglyceride lipase Yju3p. Biochim. Biophys. Acta. 2016, 1861, 462–470. [Google Scholar] [CrossRef]
- Bertrand, T.; Auge, F.; Houtmann, J.; Rak, A.; Vallee, F.; Mikol, V.; Berne, P.F.; Michot, N.; Cheuret, D.; Hoornaert, C.; et al. Structural basis for human monoglyceride lipase inhibition. J. Mol. Biol. 2010, 396, 663–673. [Google Scholar] [CrossRef]
- Rengachari, S.; Bezerra, G.A.; Riegler-Berket, L.; Gruber, C.C.; Sturm, C.; Taschler, U.; Boeszoermenyi, A.; Dreveny, I.; Zimmermann, R.; Gruber, K.; et al. The structure of monoacylglycerol lipase from Bacillus sp. H257 reveals unexpected conservation of the cap architecture between bacterial and human enzymes. Biochim. Biophys. Acta. 2012, 1821, 1012–1021. [Google Scholar] [CrossRef]
- Derewenda, U.; Brzozowski, A.M.; Lawson, D.M.; Derewenda, Z.S. Catalysis at the interface the anatomy of a conformational change in a triglyceride lipase. Biochemistry 1992, 31, 1532–1541. [Google Scholar] [CrossRef]
- Brzozowski, A.M.; Derewenda, U.; Derewenda, Z.S.; Dodson, G.G.; Lawson, D.M.; Turkenburg, J.P.; Bjorkling, F.; Huge-Jensen, B.; Patkar, S.A.; Thim, L. A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex. Nature 1991, 351, 491–494. [Google Scholar] [CrossRef]
- Gurung, N.; Ray, S.; Bose, S.; Rai, V. A broader view: Microbial enzymes and their relevance in industries, medicine, and beyond. BioMed. Res. Int. 2013, 2013, 329121. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, K.S.; Poljak, A.; Guilhaus, M.; De Francisci, D.; Curmi, P.M.; Feller, G.; D’Amico, S.; Gerday, C.; Uversky, V.N.; Cavicchioli, R. Role of lysine versus arginine in enzyme cold-adaptation: Modifying lysine to homo-arginine stabilizes the cold-adapted α-amylase from Pseudoalteramonas haloplanktis. Proteins 2006, 64, 486–501. [Google Scholar] [CrossRef]
- Trincone, A. Marine biocatalysts: Enzymatic features and applications. Marine Drugs 2011, 9, 478–499. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Lan, D.; Xin, R.; Yang, B.; Wang, Y. Screening and characterization of a thermostable lipase from marine, Streptomyces, sp. strain W007. Biotechnol. Appl. Biochem. 2016, 63, 41–50. [Google Scholar] [CrossRef]
- Wang, X.; Li, D.; Qu, M.; Durrani, R.; Yang, B.; Wang, Y. Immobilized MAS1 lipase showed high esterification activity in the production of triacylglycerols with n-3 polyunsaturated fatty acids. Food Chem. 2017, 216, 260–267. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, D.; Wang, W.; Yang, B.; Wang, Y. A highly efficient immobilized MAS1 lipase for the glycerolysis reaction of n-3 PUFA-rich ethyl esters. J. Mol. Catal. B 2016, 134, 25–31. [Google Scholar] [CrossRef]
- Wissuwa, J.; Stokke, R.; Fedoy, A.E.; Lian, K.; Smalas, A.O.; Steen, I.H. Isolation and complete genome sequence of the thermophilic Geobacillus sp. 12AMOR1 from an Arctic deep-sea hydrothermal vent site. Stand Genom. Sci. 2016, 11, 16. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Wang, W.; Durrani, R.; Li, X.; Yang, B.; Wang, Y. Simplified Enzymatic Upgrading of High-Acid Rice Bran Oil Using Ethanol as a Novel Acyl Acceptor. J. Agric. Food Chem. 2016, 64, 6730–6737. [Google Scholar] [CrossRef]
- Mogensen, J.E.; Sehgal, P.; Otzen, D.E. Activation, inhibition, and destabilization of Thermomyces lanuginosus lipase by detergents. Biochemistry 2005, 44, 1719–1730. [Google Scholar] [CrossRef]
- Yuan, D.; Lan, D.; Xin, R.; Yang, B.; Wang, Y. Biochemical properties of a new cold-active mono- and diacylglycerol lipase from marine member Janibacter sp. strain HTCC2649. Int. J. Mol. Sci. 2014, 15, 10554–10566. [Google Scholar] [CrossRef] [PubMed]
- Selvin, J.; Kennedy, J.; Lejon, D.P.; Kiran, G.S.; Dobson, A.D. Isolation identification and biochemical characterization of a novel halo-tolerant lipase from the metagenome of the marine sponge Haliclona simulans. Microb. Cell Fact. 2012, 11, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheng, J.; Wang, F.; Wang, H.; Sun, M. Cloning, characterization and expression of a novel lipase gene from marine psychrotrophic Yarrowia lipolytica. Ann. Microbiol. 2012, 62, 1071–1077. [Google Scholar] [CrossRef]
- Parra, L.P.; Reyes, F.; Acevedo, J.P.; Salazar, O.; Andrews, B.A.; Asenjo, J.A. Cloning and fusion expression of a cold-active lipase from marine Antarctic origin. Enzym. Microb. Technol. 2008, 42, 371–377. [Google Scholar] [CrossRef]
- Zhang, J.-W.; Zeng, R.-Y. Molecular cloning and expression of a cold-adapted lipase gene from an Antarctic deep sea psychrotrophic bacterium Pseudomonas sp. 7323. Mar. Biotechnol. 2008, 10, 612–621. [Google Scholar] [CrossRef] [PubMed]
- Hårdeman, F.; Sjöling, S. Metagenomic approach for the isolation of a novel low-temperature-active lipase from uncultured bacteria of marine sediment. FEMS Microbiol. Ecol. 2007, 59, 524–534. [Google Scholar] [CrossRef] [Green Version]
- Khan, F.I.; Lan, D.; Durrani, R.; Huan, W.; Zhao, Z.; Wang, Y. The Lid Domain in Lipases: Structural and Functional Determinant of Enzymatic Properties. Front. Bioeng. Biotechnol. 2017, 5, 16. [Google Scholar] [CrossRef]
- Wang, Y.; Ryu, B.H.; Yoo, W.; Lee, C.W.; Kim, K.K.; Lee, J.H.; Kim, T.D. Identification, characterization, immobilization, and mutational analysis of a novel acetylesterase with industrial potential (LaAcE) from Lactobacillus acidophilus. Biochim. Biophys. Acta. Gen. Subj. 2018, 1862, 197–210. [Google Scholar] [CrossRef]
- Jing, F.; Zhao, L.; Yandeau-Nelson, M.D.; Nikolau, B.J. Two distinct domains contribute to the substrate acyl chain length selectivity of plant acyl-ACP thioesterase. Nat. Commun. 2018, 9, 860. [Google Scholar] [CrossRef]
- Storici, F.; Lewis, L.K.; Resnick, M.A. In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 2001, 19, 773. [Google Scholar] [CrossRef]
- Kitaura, S.; Suzuki, K.; Imamura, S. Monoacylglycerol lipase from moderately thermophilic Bacillus sp. strain H-257: Molecular cloning, sequencing, and expression in Escherichia coli of the gene. J. Biochem. 2001, 129, 397–402. [Google Scholar] [CrossRef]
- Imamura, S.; Kitaura, S. Purification and characterization of a monoacylglycerol lipase from the moderately thermophilic bacillus sp. H-257. J. Biochem. 2000, 127, 419–425. [Google Scholar] [CrossRef] [PubMed]
- Morris, G.M.; Goodsell, D.S.; Halliday, R.S.; Huey, R.; Hart, W.E.; Belew, R.K.; Olson, A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998, 19, 1639–1662. [Google Scholar] [CrossRef] [Green Version]
- Bordoli, L.; Kiefer, F.; Arnold, K.; Benkert, P.; Battey, J.; Schwede, T. Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 2009, 4, 1–13. [Google Scholar] [CrossRef] [PubMed]









| Fraction | Total Activity (U) | Total Protein (mg) | Specific Activity (U/mg) | Purification Fold | Yield (%) |
|---|---|---|---|---|---|
| Lysate | 178,080 | 1484.8 | 120 | 1 | 100 |
| Affinity chromatography | 163,812 | 264 | 620.5 | 4.85 | 17.7 |
| Entry | Species | Enzyme | Substrate | pH | Temperature (°C) | Thermostability (Half-Life) | Reference |
|---|---|---|---|---|---|---|---|
| 1 | Geobacillus sp. 12AMOR1 | GMGL | Glycerin monostearate | 8.0 | 60 | 60 min at 70 °C | This work |
| 2 | Streptomyces sp. strain W007 | MAS1 | Camellia oil | 7.0 | 40 | 90 min at 70 °C | [15] |
| 3 | Janibacter sp. strain HTCC2649 | MAJ1 | DAG and MAG in Camellia oil | 7.0 | 30 | less than 30 min at 40 °C | [21] |
| 4 | Haliclona simulans | Lpc53E1 | pNP-ester | 7.0 | 40 | 60 min at 90 °C | [22] |
| 5 | Yarrowia lipolytica yeast | LIPY | pNP-ester | 8.5 | 35 | more than 120 min at 70 °C | [23] |
| 6 | Psychrobacter sp. | MBP-lipase | pNP-ester | 9 | 20 | 10 min at 40 °C | [24] |
| 7 | Pseudomonas sp. 7323 | rLipA. | pNP-ester | 9 | 30 | 4.5 h at 30 °C | [25] |
| 8 | Uncultured bacteria of marine sediment | h1Lip1 | pNP-ester | 7.25 | 35 | less than 5 min at 40 °C | [26] |
| Ester | WT | Leu142Ala | Ile145Ala | Ile170Phe | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Kinetic Parameter | Km (mM) | kcat (S−1) | kcat/Km (mM−1·S−1) | Km (mM) | kcat (S−1) | kcat/Km (mM−1·S−1) | Km (mM) | kcat (S−1) | kcat/Km (mM−1·S−1) | Km (mM) | kcat (S−1) | kcat/Km (mM−1·S−1) |
| C4 | 6.80 | 9.74 | 1.43 | 14.78 | 15.89 | 1.08 | 9.56 | 3.25 | 0.34 | 2.37 | 3.31 | 1.21 |
| C6 | 4.06 | 4.31 | 1.06 | 1.66 | 4.08 | 2.46 | 1.34 | 2.02 | 1.51 | 1.60 | 3.67 | 2.29 |
| C8 | 2.83 | 2.58 | 0.91 | 4.65 | 1.29 | 0.28 | 8.32 | 0.95 | 0.11 | 4.63 | 2.08 | 0.45 |
| Mutations | Forward/Reverse Primers |
|---|---|
| Leu142A | F: GATCTGCCGCGTTTCGCCGATGCAATCGGTTC R: GAACCGATTGCATCGGCGAAACGCGGCAGATC |
| Ile145Ala | F: CCGCGTTTCCTGGATGCAGCAGGTTCCGATATCAAAAAAC R: GTTTTTTGATATCGGAACCTGCTGCATCCAGGAAACGCGG |
| Ile170Phe | F: CAAGCATCCGTCAGTTCGTTCAGCTGATG R: CATCAGCTGAACGAACTGACGGATGCTTG |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, W.; Lan, D.; Zhao, Z.; Li, S.; Li, X.; Wang, Y. A Thermostable Monoacylglycerol Lipase from Marine Geobacillus sp. 12AMOR1: Biochemical Characterization and Mutagenesis Study. Int. J. Mol. Sci. 2019, 20, 780. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20030780
Tang W, Lan D, Zhao Z, Li S, Li X, Wang Y. A Thermostable Monoacylglycerol Lipase from Marine Geobacillus sp. 12AMOR1: Biochemical Characterization and Mutagenesis Study. International Journal of Molecular Sciences. 2019; 20(3):780. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20030780
Chicago/Turabian StyleTang, Wei, Dongming Lan, Zexin Zhao, Shuang Li, Xiuting Li, and Yonghua Wang. 2019. "A Thermostable Monoacylglycerol Lipase from Marine Geobacillus sp. 12AMOR1: Biochemical Characterization and Mutagenesis Study" International Journal of Molecular Sciences 20, no. 3: 780. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms20030780