1. Introduction
Human epidermal growth factor (hEGF), a 53-amino acid peptide with three disulfide bonds, promotes the proliferation and differentiation of epithelial cells [
1,
2,
3]. hEGF is specially recognized by membrane-localized receptor, hEGF receptor (EGFR) in human and initiates a series of intracellular signalling cascades, including the synthesis of proteins, the improvement of DNA topoisomerase activity and the activation of key genes involved in cell proliferation, differentiation, and migration [
4,
5]. Moreover, hEGF has been applied to many disease treatments based on the characteristic of promoting the growth of epidermal cells. However, owing to the high production cost of hEGF, treatment with the growth factor is limited to chronic diabetic ulcers.
The mass production of hEGF mainly contains three most common methods. Firstly, the isolation of hEGF from batches of fresh human urine, blood, breast milk, and gastric juice has been largely reported [
6]. However, endogenous concentration of hEGF is very low (50 ng/mL), and most of hEGF forms complex with receptor EGFR in vivo. The isolation and purification of hEGF in gram quantities has become a formidable task [
6]. In addition, hEGF with three disulfide bridges cannot be produced as soluble, active, and correctly folded protein via chemical synthesis method. Thirdly, producing heterologous proteins like hEGF from prokaryotic and eukaryotic organisms in large quantities based on recombinant DNA technology placed increased impetus on the development of more effective and economical methods for industrial purposes [
7].
hEGF has been expressed in various prokaryotic and eukaryotic systems, including
Escherichia coli, yeast, rice, and tobacco [
8,
9,
10,
11]. Furthermore, the bioactive hEGF has been reported to be highly expressed in tobacco hairy roots, thus demonstrating the feasibility of expressing hEGF protein in the hairy root production system [
12]. As production systems, hairy root cultures have a number of advantages, such as rapid growth rate, high productivity, genetic/biochemical stability, and hormone-autotrophy [
13]. Therefore, the hairy root systems have been widely used to produce recombinant pharmaceutical proteins, such as human acetylcholinesterase, interleukin 12, and human tissue-plasminogen activator [
14,
15,
16]. The application of hairy root cultures to the pharmaceutical industry is believed to be of great potential because of its high efficiency and relatively low cost [
17].
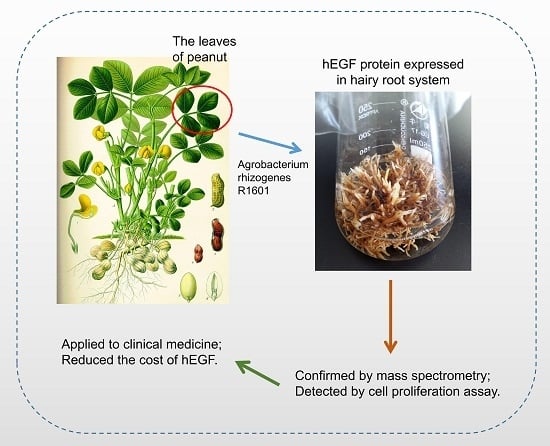
As an excellent feed used in animal husbandry, peanut contains no hazardous substances and, thus, may be used to isolate and purify hEGF safely. In the previous studies, Parsons described the properties of the peanut hairy root system, and concluded that this system offered new possibilities for expressing exogenous proteins [
12]. To construct the expression system of
hEGF in peanut hairy roots,
hEGF was inserted into pCAMBIA1300 vector, and the recombined vector was transformed into
Agrobacterium rhizogenes R1601 to obtain the engineering strain (designated as HR1601). Peanut leaves were infected by HR1601, and the
hEGF-overexpressing (OE) hairy root lines were induced and developed at the inoculation site. The protein purification and mass spectrometry assays showed that the protein expressed in peanut hairy roots was target protein hEGF. Moreover, the hEGF was demonstrated to be bioactive and non-toxic on human cells. Taken together, this study gives strong evidence that the peanut hairy roots system is suitable for the production of hEGF protein.
3. Discussion
hEGF has been widely reported to promote proliferation of a wide range of cells, such as epithelial cells, endothelial cells and fibroblast cells, which can be used as a perfect cut healing agent for various wound healing and skin regeneration [
18]. Therefore, the clinical application value or market demand of hEGF has become huge and booming in recent years. Hairy root is a good host for mass production of recombinant protein due to its abundant quality and extensive lateral branches [
19]. Hairy root production system has received considerable interest in recent years [
20,
21]. As promising systems for protein production, hairy root cultures have been used to obtain vaccine, antibody, and multiple clinical proteins [
22,
23]. As an important food crop with low potential risk, peanut hairy root system is consistently capable of producing recombinant proteins in a more native, massive and lower risk pattern than those of other plant production system [
11,
14,
24]. This study successfully expressed and purified hEGF in peanut hairy roots, and the maximum hEGF protein content in OE-3 line was 10.7 µg/g, indicating that peanut hairy root system is efficient to produce this commercial protein. Taken together, peanut hairy roots may provide a novel potential platform for the production of a variety of human or animal proteins, at least hEGF.
Notably, the growth rate of overexpressed hEGF hairy roots was lower than that of the normal control group (
Figure 2C and
Figure 3A), which can be explained by the fact that the continuous high expression of exogenous hEGF protein may restrict the expression of other proteins in peanuts in some unknown mechanism. As the positive regulator for cell growth in human, hEGF lacks the specific receptor(s) in peanut hairy roots, thus it cannot exert the role in promoting cell growth in peanut root system.
Peanuts as one of the five most important oilseed crops, serves as a good source of proteins, calories, vitamins and minerals. Thus, these food crops have low potential risk to contain hazardous substances for people and animals. Previous study exhibited that opines, resveratrol and stilbene derivatives were detected in peanut hairy roots [
20]. In our study, resveratrol contents in different transgenic lines were examined and the results showed that transgenic lines OE-1, OE-2, OE-3 and OE-4 had very similar contents of resveratrol (
Supplementary Figure S2), while the protein levels of hEGF were diverse in these transgenic lines (
Figure 2B). Therefore, our study indicates that resveratrol does not interfere with the expression of hEGF in peanuts. In addition, the molecular sieve purification system can eliminate most of hazardous substances and obtain the highly purified hEGF ultimately.
Conversion of the hEGF protein into extracellular protein instead of cytoplasmic protein by modification of the expression construct and sub-cellular targeting will be attempted in future work. Furthermore, optimizing the culture medium or other measures in appropriate method may remarkably improve the expression levels of hEGF in peanut hairy root system. Taken together, all these results demonstrate the capacity of peanut hairy root cultures as a controlled, sustainable and scalable production system that can be induced to produce valued human proteins, like hEGF.
Plant hairy root system has been favored by researchers in recent years and various studies have emerged implicating hairy root system to be widely applied in clinical field. Clinical safety and protein expression ability are two pivotal problems in clinical field.
Brassica rapa hairy root has been established in 2019, and human alpha-L-iduronidase can undergo the removal of N-terminal signal peptide and
O-glycosylation modification in the expression system, thus demonstrating the feasibility of expressing heterologous protein in the hairy root production system [
23]. In addition, hEGF produced in our study can promote human liver cells (HL-7702 cells) grown via cell proliferation assay (
Figure 4), suggesting that the hEGF produced in peanut hairy root system has biological activity. Thus, we make an assumption that the bioactive hEGF may also undergo correct post-translational modification process, including the formation of intramolecular disulfide bonds and
O-fucosylation modification process in peanuts.
Brassica rapa and peanuts as food crops are good hosts for mass production of recombinant protein. The activated hEGF has been reported to be highly expressed in tobacco hairy roots (2 μg/g, fresh weight) [
12], which is slightly higher than the peanut hairy roots system (10.7 μg/g, dry weight) in our study. However, harmful substances such as nicotine in the tobacco system has restricted the wide application of tobacco hairy root system in the clinical field. The development of molecular sieve purification system can lay a solid foundation for further exploitation of the system in the future.
4. Materials and Methods
4.1. Plant Material and Bacteria Strains
The pCAMBIA1300 expression vector used in this research was stored in Chengchao Zheng’s laboratory at the State Key Laboratory of Crop Biology, Shandong Agricultural University (Tai-an, Shandong, China). The peanut seeds (Luhua 11), Escherichia coli DH5α, and Agrobacterium rhizogenes R1601 were stored at the Gene Engineering Laboratory of the College of Pharmacy, Binzhou Medical University (Yan-tai, Shandong, China). The peanut seeds were stored at room temperature after drying and reproduced once a year. The strains of Escherichia coli DH5α and Agrobacterium rhizogenes R1601 were stored at −80 °C and activated once a year.
4.2. The Construction of HR1601 Strain
The
hEGF gene sequence was 165 base pair (bp) long and synthesized by Sangon (Shanghai, China) with the reference to the sequence demonstrated by Yamagata in 1989 [
25]. The primers for specific amplification of
hEGF were designed as follows: forward primer, GGA TCC ATG AAC AGT GAT TCA GAA TGT CCT C (
BamHI) and reverse primer, GAG CTC CTA TCG CAG TTC CCA CCA TTT C (
SalI). The PCR product was sub-cloned into pCAMBIA1300 expression vector with recognition sites for
BamHI and
SalI. Then, the expression vector was transformed into
Agrobacterium rhizogenes R1601 competent cells, and the
Agrobacterium rhizogenes R1601 with expression vector was designated as HR1601. The restriction enzymes,
BamHI and
SacI, were purchased from Takara Biotech (Dalian, China). DNA polymerase (2 × Phanta Max Master Mix, p515) was purchased from Vazyme (Nanjing, China). Trans DNA Marker and EasyPure
® Quick Gel Extraction Kit were purchased from TransGen (Beijing, China).
4.3. The Establishment of Peanut Hairy Root Culture System
Seeds of peanuts were germinated on soil and grown under greenhouse conditions (25 °C and approximately 16 h light /8 h dark photoperiod) for 30 d. Surface-sterilized leaf disks (approximately 2 × 2 cm) were inoculated with various concentrations of
Agrobacterium rhizogenes R1601 and HR1601 strain (OD
600 = 0.5, 1.0, 1.5 or 2.0) for 2 min [
19]. After removal of the excess strain with distilled water, leaf disks were then transferred to Murashige and Skoog medium (MS) containing cefotaxime sodium (200 mg/L) and allowed to grow in continuous light (8800 LUX) at 25 °C for about three weeks for hairy root emergence. The materials were subsequently cultured at 25 °C (16 h light/8 h dark, 8800 LUX) for two weeks and the roots were about 2 cm length. The root elicitation rate was calculated with the following formula: the number of leaves with initiated roots/the number of leaves infected by R1601 or HR1601. Hairy roots were cut away from the leaf blade base and transferred to MS medium to culture for another two weeks, and the roots were about 6–10 cm length. Roots of maturation zone (about 1 cm) were used to extract DNA to screen transgenic hairy roots. Then, transgenic hairy roots were transferred to 100 mL of MS liquid medium to culture in large scale for another one month for the protein extraction. In this study, 35S::
GFP (pBI121-GFP expression vector) has been used in advance to test the effectiveness of this system, and the fluorescence of GFP was monitored by fluorescent inverted microscopy (Olympus, IX71, Tokyo, Japan). At least 20 individual roots were analyzed.
4.4. Hairy Root Growth and hEGF Protein Content Analyses
Seven hEGF overexpression lines were sub-cultured for one month at 25 °C in 100 mL of MS medium, with each line separated into three parallel samples, and the peanut hairy roots infected by R1601 were used as the negative control. After removal of the excess MS medium with distilled water, the root samples were used for protein concentration analysis. The total proteins of hairy roots (0.1 g, dry weight) were extracted using the Plant Total Protein Extraction Kit (Sangon, Shanghai, China). After dissolution in 10 mM phosphate buffer (Solarbio, Beijing, China), the hEGF protein concentrations were measured by ELISA kit (Sbjbio, Nanjing, China) according to the manufacturer’s instructions.
4.5. The Detection of the hEGF Protein by Matrix-Assisted Laser Desorption/Ionization Time of Flight Mass Spectrometry
The total protein extracted by Kit were filtered with Millipore Super filter (Millipore, Billerica, MA, USA), and the proteins between 3 kDa and 10 kDa were retained. The SDS-PAGE electrophoresis was subsequently performed to isolate the hEGF protein, and the protein bond approximately 6 kDa was identified by matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF-MS) (Sangon, Shanghai, China). Then hEGF was digested into short peptides by trysin that can catalyze the hydrolysis at the specific amino acids (lysine or arginine). MALDI-TOF-MS analysis showed that four sequenced short peptides could match the protein sequence of hEGF via sequence alignment analysis, which match 47% of the total protein sequence of hEGF (29–53 amino acid). The protein sequence of hEGF was obtained and downloaded from (
https://0-www-ncbi-nlm-nih-gov.brum.beds.ac.uk/nucleotide/341526?Report=genbank&log$=nuclalign&blast_rank=1&RID=DVGPY9WK016).
4.6. Digestion Before MS Analysis
In gel digestion: according to Katayama et al. [
26]. (1) Wash the gel spot twice. (2) Remove the water and de-stained the gel spot at 23 °C for 30 min. (3) Remove the de-stained solution and add dehydration solution1 for 30 min. (4) Remove the dehydration solution1 and add dehydration solution2 for 30 min. (5) Remove the dehydration solution2 and add reduction solution1 at 57 °C for 1 h. (6) Remove the reduction solution1 and add reduction solution2 at 23 °C for 30 min. (7) Remove the reduction solution2 and add imbibition solution at 23 °C for 10 min. (8) Remove the imbibition solution and add dehydration solution1 for 30 min. (9) Remove the dehydration solution1 and add dehydration solution2 for 30 min. (10) The gels were rehydrated in 10 µl digest solution for 30 min. (11) 20 µL cover solution was added for digestion 16 h at 37 °C. (12) The supernatants were transferred into another tube. (13) The gels were extracted once with 50 µL extraction buffer at 37 °C for 30 min. The peptide extracts and the supernatant of the gel spot were combined and then completely dried.
4.7. MS Analysis
(1) Samples were re-suspended with Nano-RPLC buffer A. (2) The online Nano-RPLC was employed on the Eksigent nanoLC-Ultra™ 2D System (AB SCIEX). The samples were loaded on C18 nanoLC trap column (100 µm × 3 cm, C18, 3 µm, 150 Å) and washed by Nano-RPLC Buffer A (0.1% FA, 2% ACN) at 2 μL/min for 10 min. (3) An elution gradient of 5–35% acetonitrile (0.1% formic acid) in 30 min gradient was used on an analytical ChromXP C18 column (75 μm × 15 cm, C18, 3 μm, 120 Å) with spray tip. (4) Data acquisition was performed with a Triple TOF 5600 System (AB SCIEX, USA) fitted with a Nanospray III source (AB SCIEX, Foster city, CA, USA) and a pulled quartz tip as the emitter. Data were acquired using an ion spray voltage of 2.5 kV, curtain gas of 30 PSI, nebulizer gas of 5 PSI, and an interface heater temperature of 150 °C. For information dependent acquisition (IDA), survey scans were acquired in 250 ms and as many as 35 product ion scans were collected if they exceeded a threshold of 150 counts per second (counts/s) with a 2+ to 5+ charge-state. The total cycle time was fixed to 2.5 s. A rolling collision energy setting was applied to all precursor ions for collision-induced dissociation (CID). Dynamic exclusion was set for ½ of peak width (18 s). And the precursor was then refreshed off the exclusion list. (5) Based on combined MS and MS/MS spectra, proteins were successfully identified based on 95% or higher confidence interval of their scores in the MASCOT V2.3 search engine (Matrix Science Ltd., London, UK.), using the following search parameters: Uniprot-Komagataella pastoris (Strain ATCC 28485) database. Trypsin as the digestion enzyme. Two missed cleavage site. Fixed modifications of Carbamidomethyl (C). Partial modifications of Acetyl (Protein N-term), Deamidated (NQ), Dioxidation (W), Oxidation (M). ± 10 ppm for precursor ion tolerance and ± 0.6 Da for fragment ion tolerance.
4.8. Cell Proliferation Assay
The biological activity of hEGF produced by transgenic hairy roots was analyzed by human liver cell HL-7702 cell proliferation assay described by Pan [
27]. Same quality of hairy roots (1 g, fresh weight) infected with R1601 or HR1601 (OE1, OE2 and OE3) were used to extract the total proteins. Vacuum freeze drying the total proteins, and dissolved in sterile water, and then filtered with Millipore Super filter. The proteins between 3 kDa and 10 kDa were retained. Methylthiazolyldiphenyl-tetrazolium bromide (MTT) assay was performed to test the bioactivity of hEGF via promoting proliferation of human liver cells (HL-7702 cells) grown in medium RPMI-1640 supplemented with 10% fetal bovine serum on sterile 96-well tissue culture plate at 37 °C and 5% CO
2 for 24 h. HL-7702 cells were supplemented with 10 μL hEGF (0.1 ng/μL) expressed in the hairy roots and incubated for 24 h. Then 20 μL MTT (5 mg/mL) solutions/well were added to cells and the plates were incubated for additional 4 h. The crystals were then dissolved in 100 μL dimethyl sulfoxide per well. Absorbance was recorded at a wavelength of 490 nm with a microtiter plate reader (Bio-Tek ELX800, Winooski, VT, USA). Human liver cells HL-7702 were obtained from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China).
4.9. Statistical Tests
All experiments were performed at least three independent repetitions (
n ≥ 3). One-way ANOVA Duncan’s test (
p < 0.05 for
Figure 2B;
p < 0.01 for
Figure 4) and Student’s
t-test (***
p < 0.001) in Statistical Product and Service Solutions 24 (SPSS 24, IBM, Armonk, NY, USA) were used for statistical analysis.