A Phytochemical Constituent, (E)-Methyl-Cinnamate Isolated from Alpinia katsumadai Hayata Suppresses Cell Survival, Migration, and Differentiation in Pre-Osteoblasts
Abstract
:1. Introduction
2. Results
2.1. EMC Decreases Cell Survival and Induces Morphological Changes in Pre-Osteoblasts
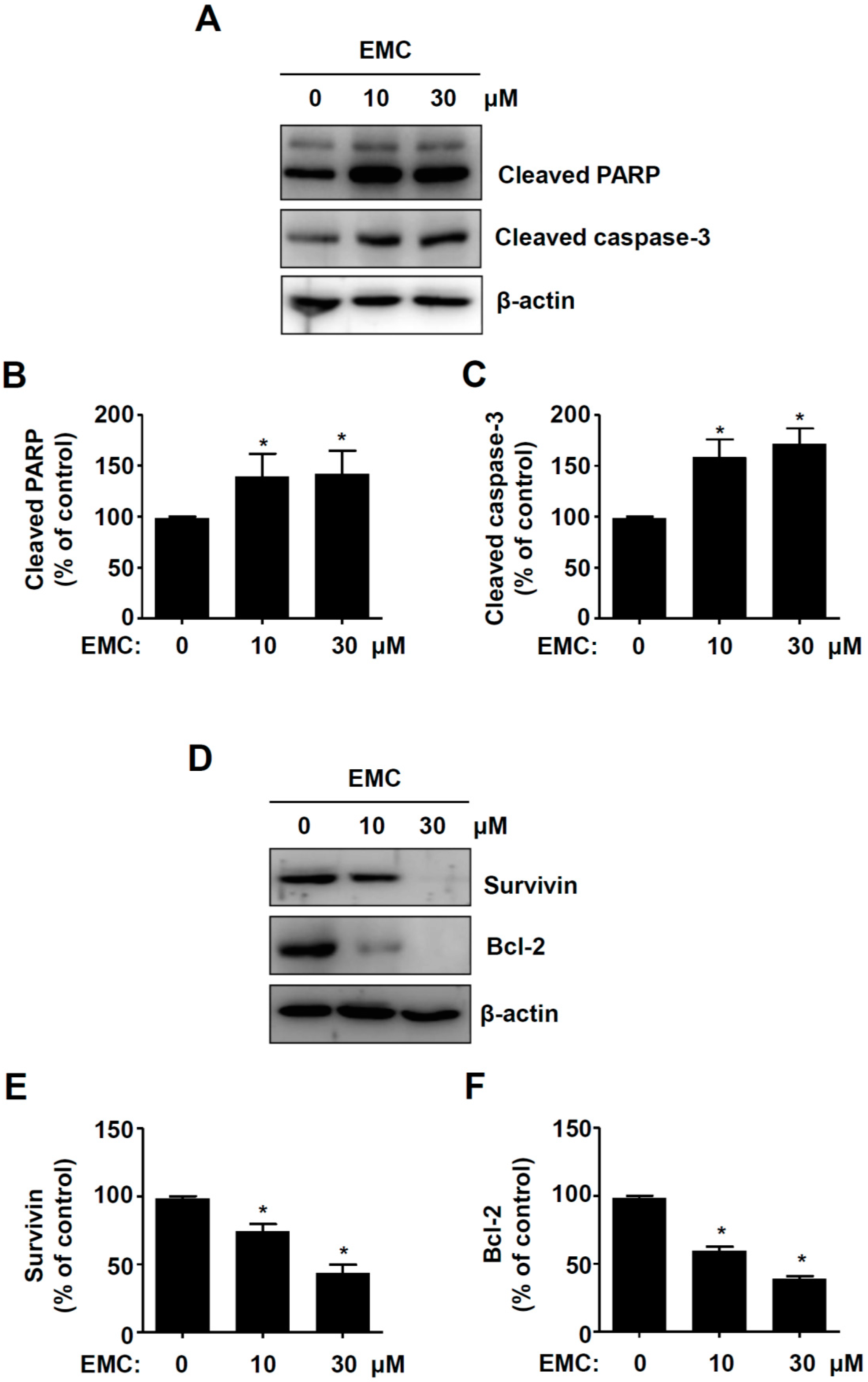
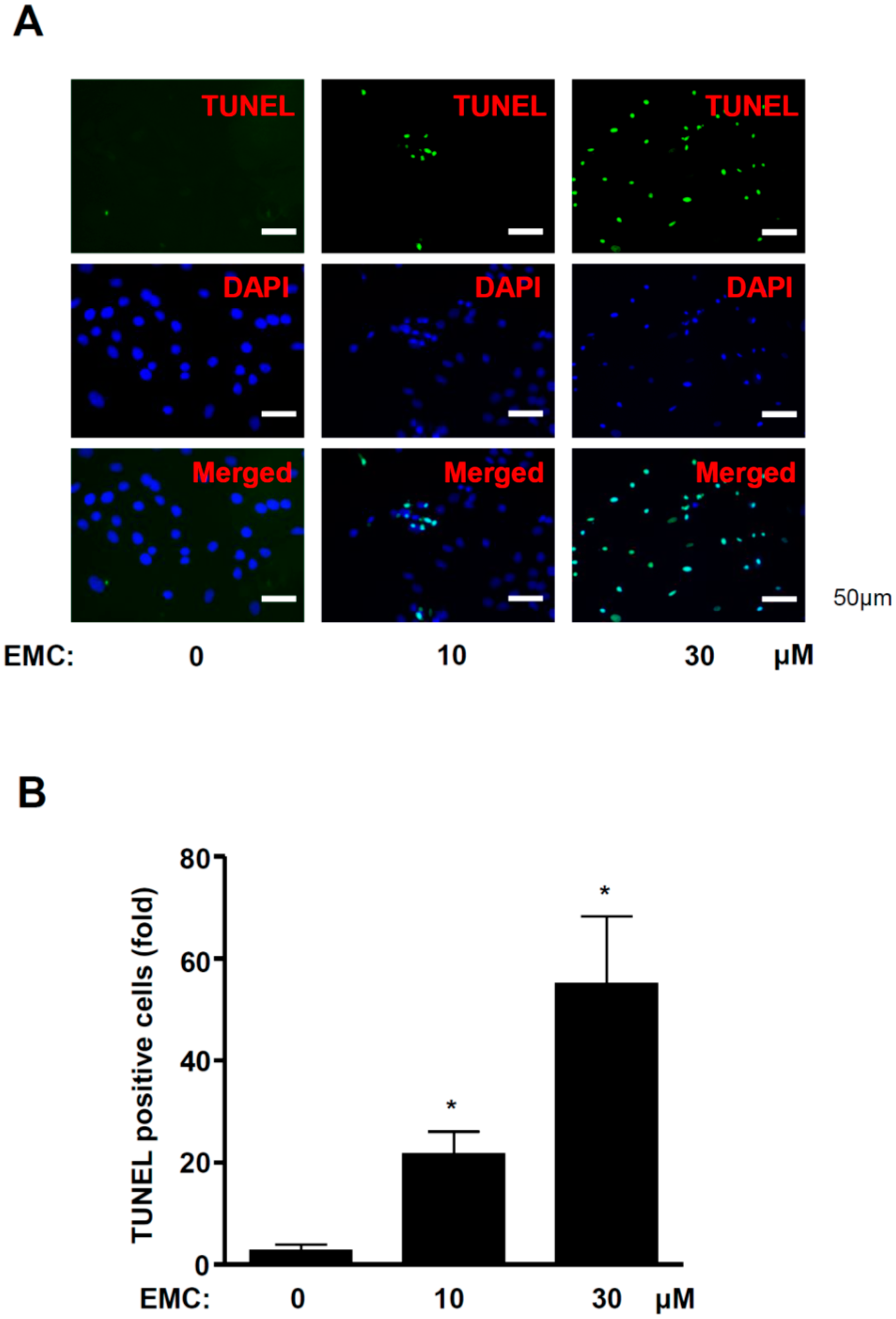
2.2. EMC Induces Apoptosis in Pre-Osteoblasts
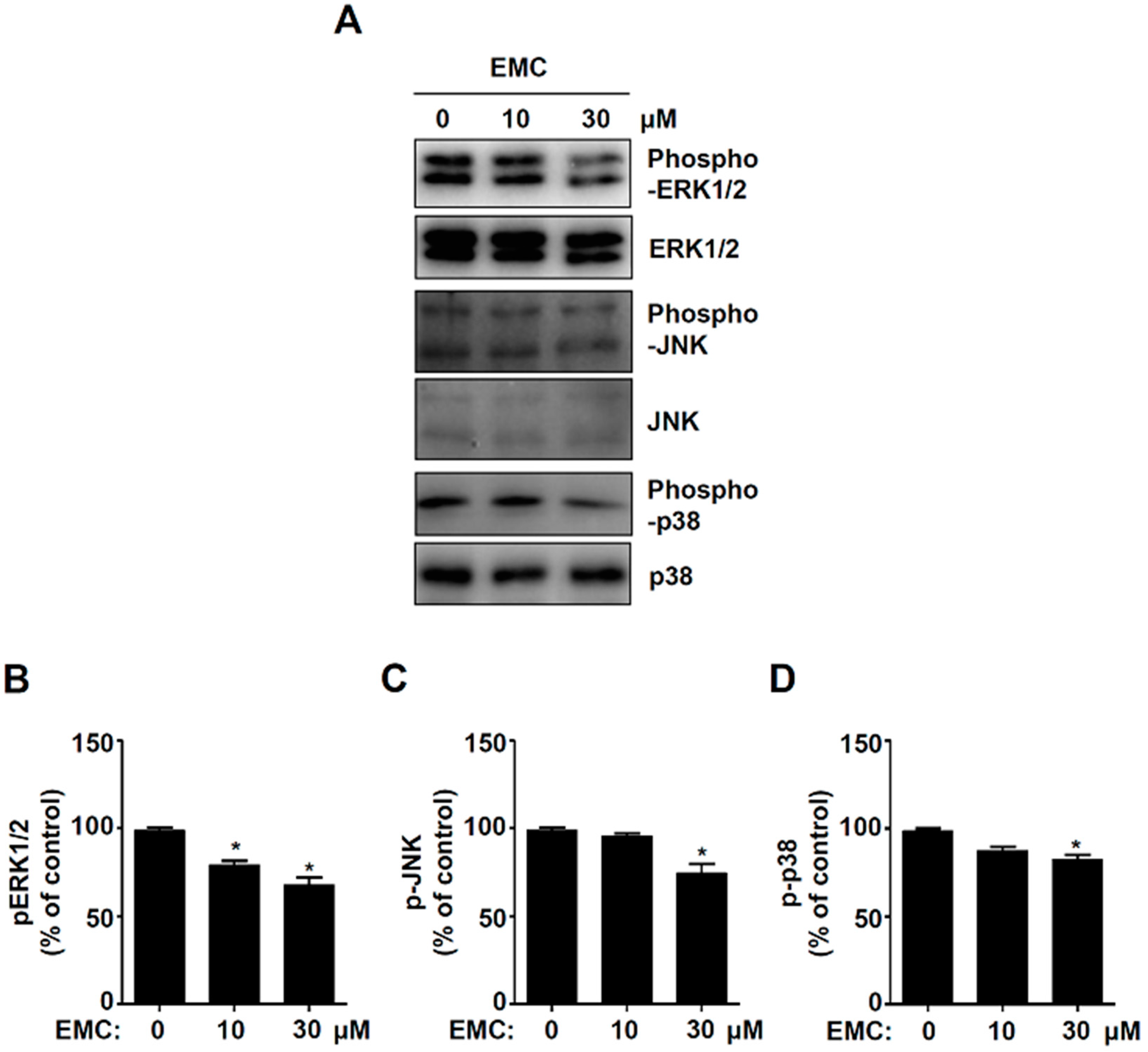
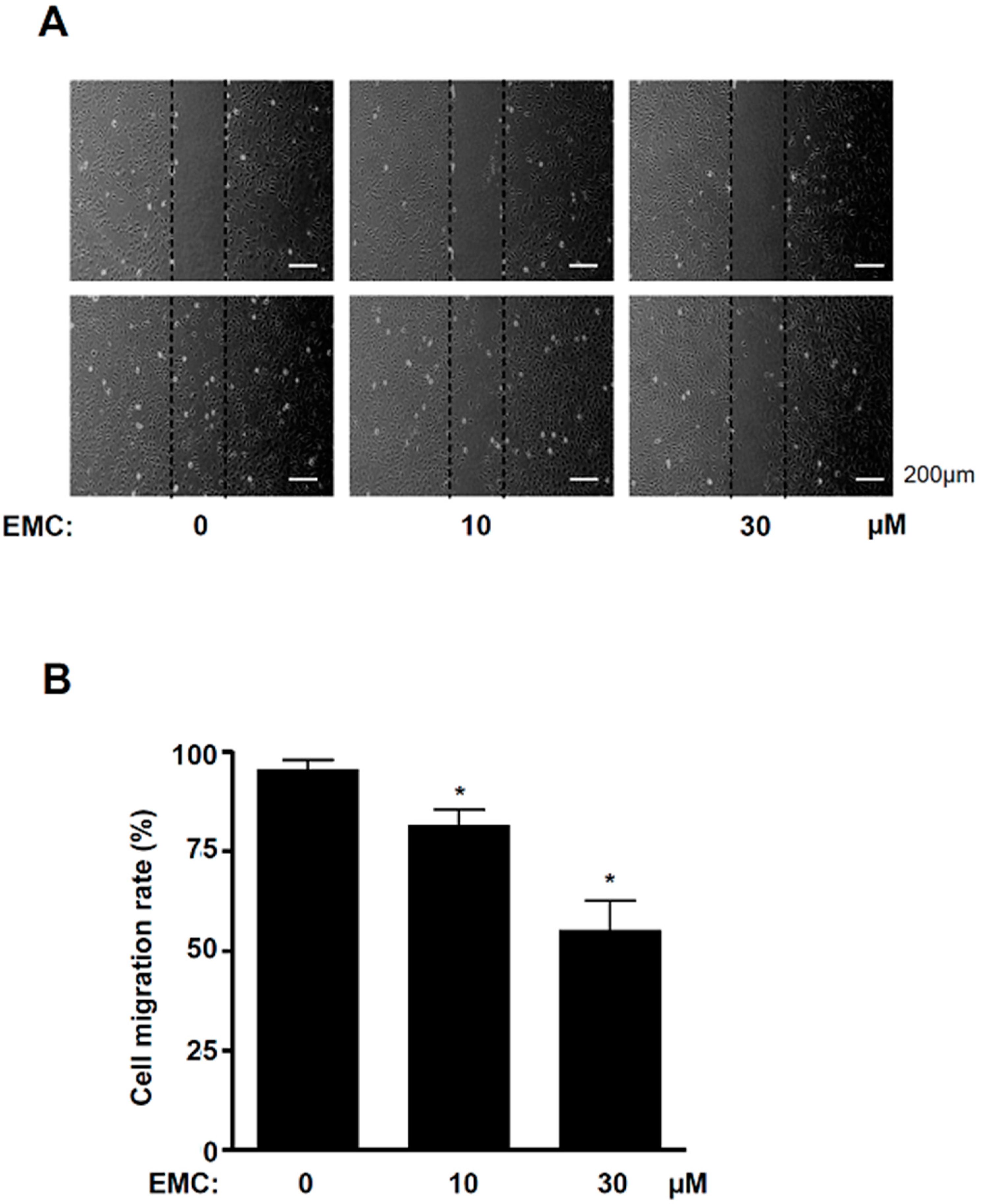
2.3. EMC Decreases MAPKs Signaling and Cell Migration in Pre-Osteoblasts
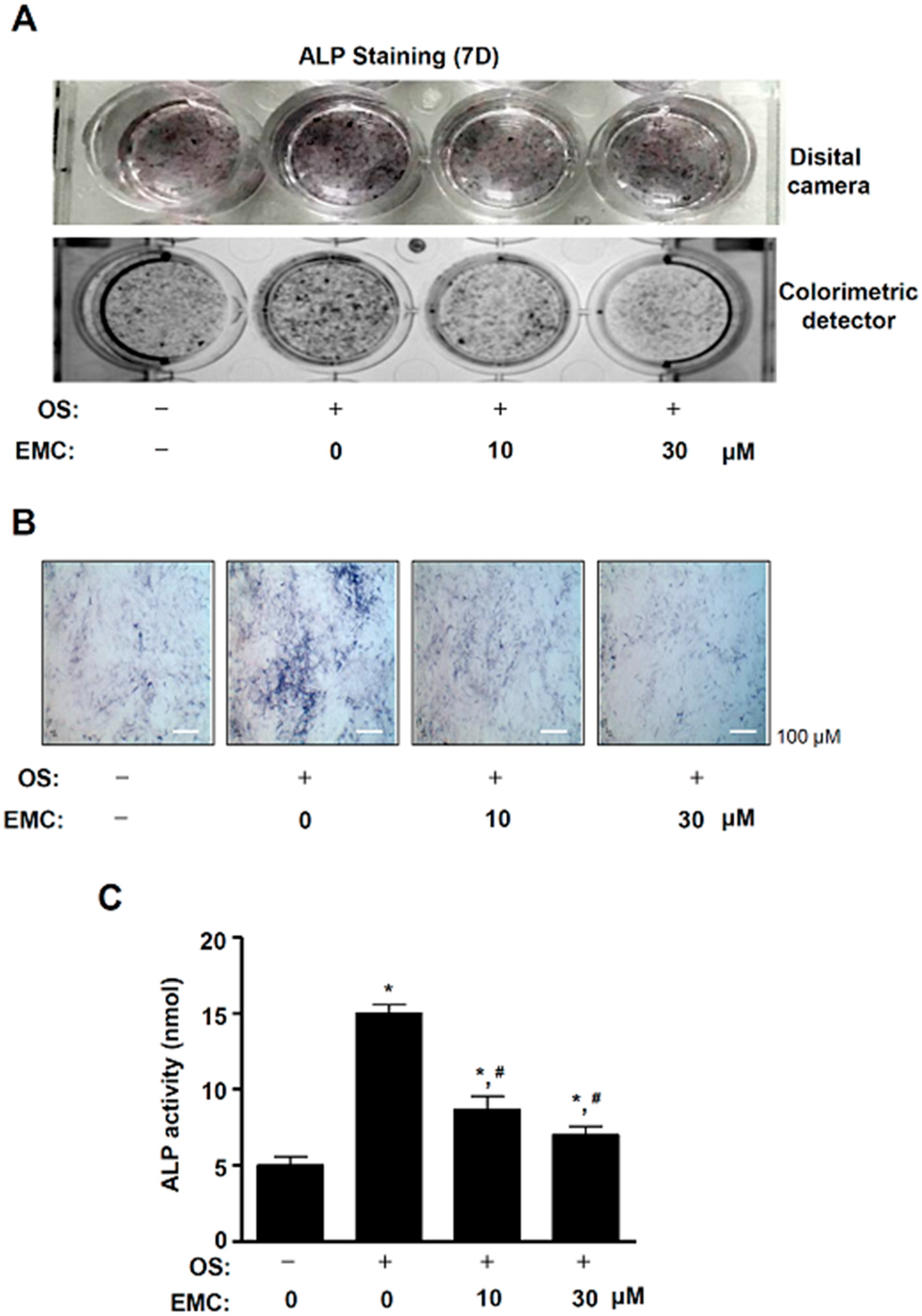
2.4. EMC Suppresses the Osteoblast Differentiation of Pre-Osteoblasts
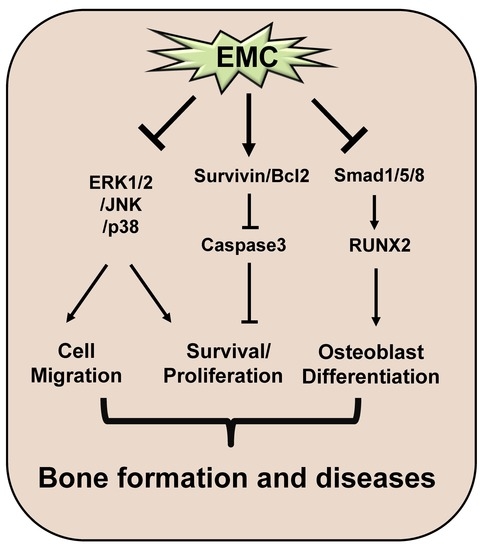
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Extraction and Isolation of Methyl-Cinnamate
4.3. (E)-Methyl-Cinnamate (EMC)
4.4. Nuclear Magnetic Resonance (NMR)
4.5. Culture of Pre-Osteoblast MC3T3-E1 Cells and Osteoblast Differentiation
4.6. MTT Assay
4.7. Western Blot Analysis
4.8. TUNEL Assay
4.9. Cell Migration Assay
4.10. ALP Staining Assay
4.11. ALP Activity Assay
4.12. Real-Time PCR Analysis
4.13. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ALP | Alkaline phosphatase |
| β-GP | β -glycerophosphate |
| EMC | (E)-methyl-cinnamate |
| L-AA | L-ascorbic acid |
| MAPKs | Mitogen-activated protein kinases |
| MSCs | Mesenchymal stem cells |
| MTT | 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) |
| OS | Osteogenic supplement |
| PARP | Poly (ADP-ribose) polymerase |
| RUNX2 | Runt-related transcription factor 2 |
References
- Lee, S.E.; Shin, H.T.; Hwang, H.J.; Kim, J.H. Antioxidant activity of extracts from Alpinia katsumadai seed. Phytother. Res. 2003, 17, 1041–1047. [Google Scholar] [CrossRef] [PubMed]
- Nam, J.W.; Seo, E.K. Structural characterization and biological effects of constituents of the seeds of Alpinia katsumadai (Alpina Katsumadai Seed). Nat. Prod. Commun. 2012, 7, 795–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, W.; Yao, L.; He, X.; Wang, L.; Li, M.; Yang, Y.; Wan, C. Hypoglycemic activity and constituents analysis of blueberry (Vaccinium corymbosum) fruit extracts. Diabetes Metab. Syndr. Obes. 2018, 11, 357–366. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Chen, X.; Crandall-Stotler, B.; Qian, P.; Kollner, T.G.; Guo, H.; Chen, F. Biosynthesis of methyl (E)-cinnamate in the liverwort Conocephalum salebrosum and evolution of cinnamic acid methyltransferase. Phytochemistry 2019, 164, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, S.P.; Wellington, G.A.; Cocchiara, J.; Lalko, J.; Letizia, C.S.; Api, A.M. Fragrance material review on methyl cinnamate. Food Chem. Toxicol. 2007, 45, S113–S197. [Google Scholar] [CrossRef] [PubMed]
- Shimoi, K.; Nakamura, Y.; Noro, T.; Tomita, I.; Fukushima, S.; Inoue, T.; Kada, T. Methyl cinnamate derivatives enhance UV-induced mutagenesis due to the inhibition of DNA excision repair in Escherichia coli B/r. Mutat. Res. 1985, 146, 15–22. [Google Scholar] [CrossRef]
- Huang, Q.S.; Zhu, Y.J.; Li, H.L.; Zhuang, J.X.; Zhang, C.L.; Zhou, J.J.; Li, W.G.; Chen, Q.X. Inhibitory effects of methyl trans-cinnamate on mushroom tyrosinase and its antimicrobial activities. J. Agric. Food Chem. 2009, 57, 2565–2569. [Google Scholar] [CrossRef]
- Lima, F.J.; Cosker, F.; Brito, T.S.; Ribeiro-Filho, H.V.; Silva, C.M.; Aragao, K.S.; Lahlou, S.; Souza, M.H.; Santos, A.A.; Magalhaes, P.J. Antispasmodic and myorelaxant effects of the flavoring agent methyl cinnamate in gut: Potential inhibition of tyrosine kinase. Eur. J. Pharmacol. 2014, 740, 192–199. [Google Scholar] [CrossRef]
- Gui, Y.; Chen, L.; Duan, S.; Li, G.; Tang, J.; Li, A. Methyl cinnamate alleviated CCI-induced upregualtion of spinal AMPA receptors and pain hypersensitivity by targeting AMPK. Eur. J. Pharmacol. 2018, 833, 183–189. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Lee, M.H.; Hsu, C.C.; Wei, C.L.; Tsai, Y.C. Methyl cinnamate inhibits adipocyte differentiation via activation of the CaMKK2-AMPK pathway in 3T3-L1 preadipocytes. J. Agric. Food Chem. 2012, 60, 955–963. [Google Scholar] [CrossRef]
- Fakhry, M.; Hamade, E.; Badran, B.; Buchet, R.; Magne, D. Molecular mechanisms of mesenchymal stem cell differentiation towards osteoblasts. World J. Stem Cells 2013, 5, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Bonewald, L.F. The amazing osteocyte. J. Bone Miner. Res. 2011, 26, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Hughes, D.E.; Boyce, B.F. Apoptosis in bone physiology and disease. Mol. Pathol. 1997, 50, 132–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iguchi, M.; Hiroi, M.; Kanegae, H.; Ohmori, Y. Costimulation of Murine Osteoblasts with Interferon-gamma and Tumor Necrosis Factor-alpha Induces Apoptosis through Downregulation of Bcl-2 and Release of Cytochrome c from Mitochondria. Mediat. Inflamm. 2018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mollazadeh, S.; Fazly Bazzaz, B.S.; Kerachian, M.A. Role of apoptosis in pathogenesis and treatment of bone-related diseases. J. Orthop. Surg. Res. 2015, 10, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karsenty, G.; Kronenberg, H.M.; Settembre, C. Genetic control of bone formation. Annu. Rev. Cell Dev. Biol. 2009, 25, 629–648. [Google Scholar] [CrossRef]
- Zheng, X.; Dai, J.; Zhang, H.; Ge, Z. MicroRNA-221 promotes cell proliferation, migration, and differentiation by regulation of ZFPM2 in osteoblasts. Braz. J. Med. Biol. Res. 2018, 51, e7574. [Google Scholar] [CrossRef]
- Karsenty, G.; Wagner, E.F. Reaching a genetic and molecular understanding of skeletal development. Dev. Cell 2002, 2, 389–406. [Google Scholar] [CrossRef] [Green Version]
- Manolagas, S.C. Birth and death of bone cells: Basic regulatory mechanisms and implications for the pathogenesis and treatment of osteoporosis. Endocr. Rev. 2000, 21, 115–137. [Google Scholar]
- Park, K.R.; Kim, J.Y.; Kim, E.C.; Yun, H.M.; Hong, J.T. RANKL-induced osteoclastogenesis is suppressed by 4-O-methylhonokiol in bone marrow-derived macrophages. Arch. Pharm. Res. 2017, 40, 933–942. [Google Scholar] [CrossRef]
- Numan, M.S.; Amiable, N.; Brown, J.P.; Michou, L. Paget’s disease of bone: An osteoimmunological disorder? Drug Des. Dev. Ther. 2015, 9, 4695–4707. [Google Scholar]
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516. [Google Scholar] [CrossRef] [PubMed]
- Rosen, A.; Casciola-Rosen, L. Macromolecular substrates for the ICE-like proteases during apoptosis. J. Cell Biochem. 1997, 64, 50–54. [Google Scholar] [CrossRef]
- Miura, M.; Chen, X.D.; Allen, M.R.; Bi, Y.; Gronthos, S.; Seo, B.M.; Lakhani, S.; Flavell, R.A.; Feng, X.H.; Robey, P.G.; et al. A crucial role of caspase-3 in osteogenic differentiation of bone marrow stromal stem cells. J. Clin. Investig. 2004, 114, 1704–1713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwaki, A.; Jingushi, S.; Oda, Y.; Izumi, T.; Shida, J.I.; Tsuneyoshi, M.; Sugioka, Y. Localization and quantification of proliferating cells during rat fracture repair: Detection of proliferating cell nuclear antigen by immunohistochemistry. J. Bone Miner. Res. 1997, 12, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Ichida, M.; Yui, Y.; Yoshioka, K.; Tanaka, T.; Wakamatsu, T.; Yoshikawa, H.; Itoh, K. Changes in cell migration of mesenchymal cells during osteogenic differentiation. FEBS Lett. 2011, 585, 4018–4024. [Google Scholar] [CrossRef] [Green Version]
- Westhoff, M.A.; Serrels, B.; Fincham, V.J.; Frame, M.C.; Carragher, N.O. SRC-mediated phosphorylation of focal adhesion kinase couples actin and adhesion dynamics to survival signaling. Mol. Cell Biol. 2004, 24, 8113–8133. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.M.; Shih, Y.T.; Chen, Y.S.; Liu, C.L.; Fang, W.K.; Tsai, C.H.; Tsai, F.J.; Kuo, W.W.; Lai, T.Y.; Huang, C.Y. Schwann Cell Migration Induced by Earthworm Extract via Activation of PAs and MMP2/9 Mediated through ERK1/2 and p38. Evid. Based Complement. Alternat. Med. 2011, 2011. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Liu, F.C.; Chou, P.Y.; Chien, Y.C.; Chang, W.S.; Huang, G.J.; Wu, C.H.; Sheu, M.J. Ethanol extracts of fruiting bodies of Antrodia cinnamomea suppress CL1-5 human lung adenocarcinoma cells migration by inhibiting matrix metalloproteinase-2/9 through ERK, JNK, p38, and PI3K/Akt signaling pathways. Evid. Based Complement. Alternat. Med. 2012, 2011. [Google Scholar] [CrossRef] [Green Version]
- Liao, X.; Lu, S.; Zhuo, Y.; Winter, C.; Xu, W.; Wang, Y. Visualization of Src and FAK activity during the differentiation process from HMSCs to osteoblasts. PLoS ONE 2012, 7, e42709. [Google Scholar] [CrossRef]
- Orimo, H. The mechanism of mineralization and the role of alkaline phosphatase in health and disease. J. Nippon Med. Sch. 2010, 77, 4–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golub, E.E.; Harrison, G.; Taylor, A.G.; Camper, S.; Shapiro, I.M. The role of alkaline phosphatase in cartilage mineralization. Bone Miner. 1992, 17, 273–278. [Google Scholar] [CrossRef]
- Yun, H.M.; Park, K.R.; Hong, J.T.; Kim, E.C. Peripheral serotonin-mediated system suppresses bone development and regeneration via serotonin 6 G-protein-coupled receptor. Sci. Rep. 2016, 6, 30985. [Google Scholar] [CrossRef] [PubMed]
- Gaur, T.; Lengner, C.J.; Hovhannisyan, H.; Bhat, R.A.; Bodine, P.V.; Komm, B.S.; Javed, A.; van Wijnen, A.J.; Stein, J.L.; Stein, G.S.; et al. Canonical WNT signaling promotes osteogenesis by directly stimulating Runx2 gene expression. J. Biol. Chem. 2005, 280, 33132–33140. [Google Scholar] [CrossRef] [Green Version]
- Phimphilai, M.; Zhao, Z.; Boules, H.; Roca, H.; Franceschi, R.T. BMP signaling is required for RUNX2-dependent induction of the osteoblast phenotype. J. Bone Miner. Res. 2006, 21, 637–646. [Google Scholar] [CrossRef] [Green Version]
- Park, K.R.; Yun, H.M. RANKL-induced osteoclastogenesis in bone marrow-derived macrophages is suppressed by cisapride. Toxicology 2019, 422, 95–101. [Google Scholar] [CrossRef]
- Park, K.R.; Kim, E.C.; Hong, J.T.; Yun, H.M. Dysregulation of 5-hydroxytryptamine 6 receptor accelerates maturation of bone-resorbing osteoclasts and induces bone loss. Theranostics 2018, 8, 3087–3098. [Google Scholar] [CrossRef]
- Park, K.R.; Yun, H.M.; Hong, J.T. G721-0282 inhibits cell growth and induces apoptosis in human osteosarcoma through down-regulation of the STAT3 pathway. Int. J. Biol. Sci. 2020, 16, 330–341. [Google Scholar] [CrossRef] [Green Version]
- Sequeira, D.B.; Seabra, C.M.; Palma, P.J.; Cardoso, A.L.; Peca, J.; Santos, J.M. Effects of a New Bioceramic Material on Human Apical Papilla Cells. J. Funct. Biomater. 2018, 9, 74. [Google Scholar] [CrossRef] [Green Version]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, K.-R.; Lee, H.; Cho, M.; Yun, H.-M. A Phytochemical Constituent, (E)-Methyl-Cinnamate Isolated from Alpinia katsumadai Hayata Suppresses Cell Survival, Migration, and Differentiation in Pre-Osteoblasts. Int. J. Mol. Sci. 2020, 21, 3700. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21103700
Park K-R, Lee H, Cho M, Yun H-M. A Phytochemical Constituent, (E)-Methyl-Cinnamate Isolated from Alpinia katsumadai Hayata Suppresses Cell Survival, Migration, and Differentiation in Pre-Osteoblasts. International Journal of Molecular Sciences. 2020; 21(10):3700. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21103700
Chicago/Turabian StylePark, Kyung-Ran, Hanna Lee, MyoungLae Cho, and Hyung-Mun Yun. 2020. "A Phytochemical Constituent, (E)-Methyl-Cinnamate Isolated from Alpinia katsumadai Hayata Suppresses Cell Survival, Migration, and Differentiation in Pre-Osteoblasts" International Journal of Molecular Sciences 21, no. 10: 3700. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21103700