Genome Editing in Cereals: Approaches, Applications and Challenges
Abstract
:1. Introduction
2. Basic Approaches Used for Genome-Editing
2.1. Zinc Finger Nucleases
2.2. Transcription Activator-Like Effectors Nucleases
2.3. CRISPR/Cas9 Based Genome Editing
3. Precise Genome Editing in Cereals
4. Genome Editing Specificity
5. Predicted Boom over Coming Years
6. CRISPR/Cas9 Based Genome Editing in Cereals
6.1. Priority Traits
6.1.1. Resistance Against Bacterial Disease
6.1.2. Resistance against Fungal Disease
6.1.3. Resistance against Insect
6.1.4. Resistance against Viruses
6.1.5. Resistance and Tolerance against Herbicide
6.1.6. Enhanced Quality and Yield
6.2. Other Traits
7. Genome Editing for Well Characterized Genes
Genes Previously Characterized by RNAi
8. Advances in CRISPR/Cas9 Based Approaches
8.1. Multi-Target Approaches
8.1.1. Csy4 Nuclease Based Multi-Target Genome Editing
8.1.2. Polycistronic t-RNA Transcripts Based Multi Target Genome Editing
8.1.3. Drosha MiRNA Based Multi Target Genome Editing
9. In the Absence of Integration Gene Expression and DNA Transfer
9.1. T-DNA Approach
9.2. Cas9 Alternatives for More Precision
10. Preferred Promoters and Methods of Transformation for Genome-Editing of Cereals
10.1. Preferred Promoters for Gene Expression Regulation in Cereals
10.2. Constitutive Promoters
10.3. Promoters Specific for Tissue or Developmental Stage
10.4. Inducible Promoter
10.5. Synthetic Promoters
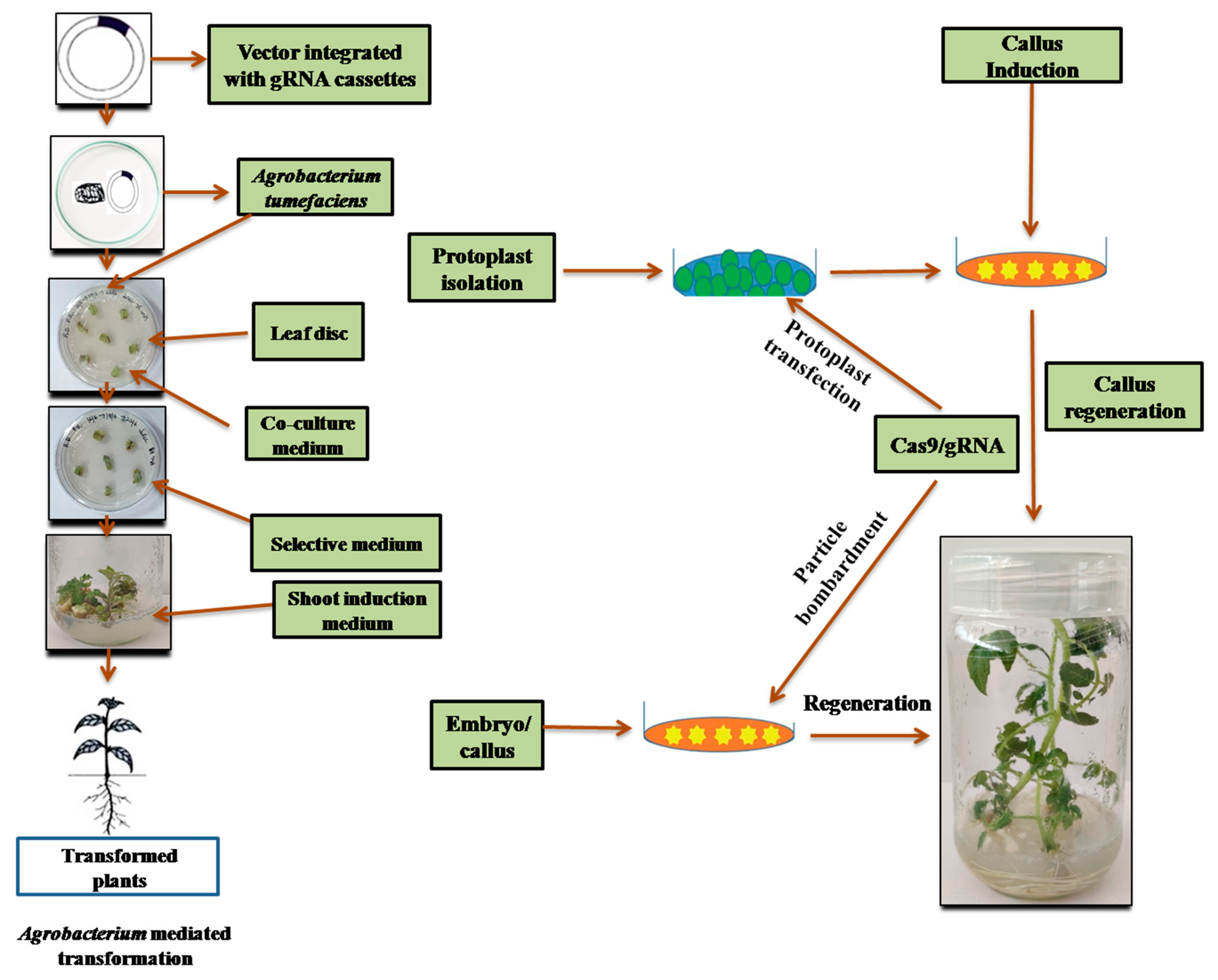
11. Transformation Methods for Genome Editing
11.1. Methods to Validate the Construct
11.1.1. Protoplast
11.1.2. Protoplast Transfection
11.1.3. Agroinfiltration Methods
11.1.4. Hairy Roots Validation
11.1.5. High Precision Base Editing
11.2. Gene Replacement
11.3. Gene Expression Modulation
12. Challenges for Genome-Editing in Cereals
12.1. Polyploidy
12.2. Transformation Efficiency
13. Off-Target Effect with Cas9 and Improved Variant of Cas9
14. Regulatory and Ethical Issues
15. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Miglani, G.S. Genome editing in crop improvement: Present scenario and future prospects. J. Crop Improv. 2017, 31, 453–559. [Google Scholar] [CrossRef]
- Weeks, D.P.; Spalding, M.H.; Yang, B. Use of designer nucleases for targeted gene and genome editing in plants. Plant Biotechnol. J. 2016, 14, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Jiang, F.; Taylor, D.W.; Sternberg, S.H.; Kaya, E.; Ma, E.; Anders, C.; Hauer, M.; Zhou, K.; Lin, S.; et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 2014, 343, 1247997. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khush, G.S. Strategies for increasing the yield potential of cereals: Case of rice as an example. Plant Breed. 2013, 132, 433–436. [Google Scholar] [CrossRef]
- Church, G.M.; Sheen, J. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar]
- Xu, R.; Yang, Y.; Qin, R.; Li, H.; Qiu, C.; Li, L.; Wei, P.; Yang, J. Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice. J. Genet. Genom. 2016, 43, 529. [Google Scholar]
- Mishra, R.; Zhao, K. Genome editing technologies and their applications in crop improvement. Plant Biotechnol. Rep. 2018, 12, 57–68. [Google Scholar] [CrossRef]
- Iqbal, M.; Raja, N.I.; Hussain, M.; Ejaz, M.; Yasmeen, F. Effect of silver nanoparticles on growth of wheat under heat stress. Iran. J. Sci. Technol. Trans. A 2019, 43, 387–395. [Google Scholar] [CrossRef]
- Svitashev, S.; Young, J.K.; Schwartz, C.; Gao, H.; Falco, S.C.; Cigan, A.M. Targeted mutagenesis, precise gene editing, and site-specific gene insertion in maize using Cas9 and guide RNA. Plant Physiol. 2015, 169, 931–945. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.; Song, N.; Sun, S.; Yang, W.; Zhao, H.; Song, W.; Lai, J. Efficiency and inheritance of targeted mutagenesis in maize using CRISPR-Cas9. J. Genet. Genom. 2016, 43, 5–36. [Google Scholar] [CrossRef]
- Feng, C.; Yuan, J.; Wang, R.; Liu, Y.; Birchler, J.A.; Han, F. Efficient targeted genome modification in maize using CRISPR/Cas9 system. J. Genet. Genom. 2016, 43, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS 8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 2017, 15, 207–216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shan, Q.; Wang, Y.; Li, J.; Gao, C. Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 2014, 9, 2395. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014, 32, 947. [Google Scholar] [CrossRef] [PubMed]
- Connorton, J.M.; Jones, E.R.; Rodríguez-Ramiro, I.; Fairweather-Tait, S.; Uauy, C.; Balk, J. Wheat vacuolar iron transporter TaVIT2 transports Fe and Mn and is effective for biofortification. Plant Physiol. 2017, 174, 2434–2444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Bai, Y.; Wu, G.; Zou, S.; Chen, Y.; Gao, C.; Tang, D. Simultaneous modification of three homoeologs of Ta EDR 1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 2017, 91, 714–724. [Google Scholar] [CrossRef] [Green Version]
- Liang, Z.; Chen, K.; Li, T.; Zhang, Y.; Wang, Y.; Zhao, Q.; Liu, J.; Zhang, H.; Liu, C.; Ran, Y.; et al. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 2017, 8, 14261. [Google Scholar] [CrossRef]
- Sánchez-León, S.; Gil-Humanes, J.; Ozuna, C.V.; Giménez, M.J.; Sousa, C.; Voytas, D.F.; Barro, F. Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol. J. 2018, 16, 902–910. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, J.; Huang, J.; Lan, H.; Wang, C.; Yin, C.; Wu, Y.; Tang, H.; Qian, Q.; Li, J.; et al. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. USA 2012, 109, 21534–21539. [Google Scholar] [CrossRef] [Green Version]
- Xie, K.; Yang, Y. RNA-guided genome editing in plants using a CRISPR–Cas system. Mol. Plant 2013, 6, 1975–1983. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; He, M.; Li, J.; Chen, L.; Huang, Z.; Zheng, S.; Zhu, L.; Ni, E.; Jiang, D.; Zhao, B.; et al. Development of commercial thermo-sensitive genic male sterile rice accelerates hybrid rice breeding using the CRISPR/Cas9-mediated TMS5 editing system. Sci. Rep. 2016, 6, 37395. [Google Scholar] [PubMed] [Green Version]
- Sun, Y.; Jiao, G.; Liu, Z.; Zhang, X.; Li, J.; Guo, X.; Du, W.; Du, J.; Francis, F.; Zhao, Y.; et al. Generation of high-amylose rice through CRISPR/Cas9-mediated targeted mutagenesis of starch branching enzymes. Front. Plant Sci. 2017, 8, 298. [Google Scholar] [PubMed]
- Li, J.; Meng, X.; Zong, Y.; Chen, K.; Zhang, H.; Liu, J.; Li, J.; Gao, C. Gene replacements and insertions in rice by intron targeting using CRISPR-Cas9. Nat. Plants 2016, 2, 16139. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, X.; Zhou, Z.; Wu, P.; Fang, M.; Pan, X.; Lin, Q.; Luo, W.; Wu, G.; Li, H. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front. Plant Sci. 2016, 7, 377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Liang, Z.; Zong, Y.; Wang, Y.; Liu, J.; Chen, K.; Qiu, J.L.; Gao, C. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016, 7, 12617. [Google Scholar]
- Wang, F.J.; Wang, C.L.; Liu, P.Q.; Lei, C.L.; Hao, W.; Gao, Y.; Liu, Y.G.; Zhao, K.J. Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the erf transcription factor gene OsERF922. PLoS ONE 2016, 11, e0154027. [Google Scholar] [CrossRef]
- Lu, Y.; Zhu, J.K. Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol. Plant 2017, 10, 523–525. [Google Scholar]
- Cordones, N.M.; Mohamed, S.; Tanoi, K.; Kobayashi, N.I.; Takagi, K.; Vernet, A.; Guiderdoni, E.; Périn, C.; Sentenac, H.; Véry, A.A. Production of low-Cs+ rice plants by inactivation of the K+ transporter Os HAK 1 with the CRISPR-Cas system. Plant J. 2017, 92, 43–56. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Zhang, X.; Wu, C.; He, Y.; Ma, Y.; Hou, H.; Guo, X.; Du, W.; Zhao, Y.; Xia, L. Engineering herbicide-resistant rice plants through CRISPR/Cas9- mediated homologous recombination of acetolactate synthase. Mol. Plant 2016, 9, 628–631. [Google Scholar]
- Tang, L.; Mao, B.; Li, Y.; Lv, Q.; Zhang, L.; Chen, C.; He, H.; Wang, W.; Zeng, X.; Shao, Y.; et al. Knockout of OsNramp5 using the CRISPR/Cas9 system produces low Cd-accumulating indica rice without compromising yield. Sci. Rep. 2017, 7, 14438. [Google Scholar]
- Macovei, A.; Sevilla, N.R.; Cantos, C.; Jonson, G.B.; Slamet-Loedin, I.; Čermák, T.; Voytas, D.F.; Choi, I.R.; Chadha-Mohanty, P. Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol. J. 2018, 16, 918–1927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, X.; Zheng, Y.; Xiao, K.; Wei, Y.; Zhu, Y.; Cai, Q.; Chen, L.; Xie, H.; Zhang, J. OsPRX2 contributes to stomatal closure and improves potassium deficiency tolerance in rice. Biochem. Biophys. Res. Commun. 2018, 495, 461–467. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhang, H.; Botella, J.R.; Zhu, J.K. Generation of new glutinous rice by CRISPR/Cas9-targeted mutagenesis of the Waxy gene in elite rice varieties. J. Integr. Plant Biol. 2018, 60, 369–375. [Google Scholar] [CrossRef] [PubMed]
- Endo, M.; Mikami, M.; Endo, A.; Kaya, H.; Itoh, T.; Nishimasu, H.; Nureki, O.; Toki, S. Genome editing in plants by engineered CRISPR–Cas9 recognizing NG PAM. Nat. Plants 2019, 5, 14. [Google Scholar]
- Saika, H.; Mori, A.; Endo, M.; Toki, S. Targeted deletion of rice retrotransposon Tos17 via CRISPR/Cas9. Plant Cell Rep. 2019, 38, 455–458. [Google Scholar]
- Chao, S.; Cai, Y.; Feng, B.; Jiao, G.; Sheng, Z.; Luo, J.; Tang, S.; Wang, J.; Hu, P.; Wei, X. Editing of rice Isoamylase Gene ISA1 provides insights into its function in starch formation. Rice Sci. 2019, 26, 77–87. [Google Scholar]
- Zhang, A.; Liu, Y.; Wang, F.; Li, T.; Chen, Z.; Kong, D.; Bi, J.; Zhang, F.; Luo, X.; Wang, J.; et al. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 2019, 39, 47. [Google Scholar]
- Jiang, M.; Liu, Y.; Liu, Y.; Tan, Y.; Huang, J.; Shu, Q. Mutation of Inositol 1, 3, 4-trisphosphate 5/6-kinase6 impairs plant growth and phytic acid synthesis in rice. Plants 2019, 8, 114. [Google Scholar] [CrossRef] [Green Version]
- Lawrenson, T.; Shorinola, O.; Stacey, N.; Li, C.; Ostergaard, L.; Patron, N.; Uauy, C.; Harwood, W. Induction of targeted, heritable mutations in barley and Brassica oleracea using RNA-guided Cas9 nuclease. Genome Biol. 2015, 16, 258. [Google Scholar] [CrossRef] [Green Version]
- Fiaz, S.; Ahmad, S.; Noor, M.A.; Wang, X.; Younas, A.; Riaz, A.; Riaz, A.; Ali, F. Applications of the CRISPR/Cas9 system for rice grain quality improvement: Perspectives and opportunities. Int. J. Mol. Sci. 2019, 20, 888. [Google Scholar]
- Shukla, V.K.; Doyon, Y.; Miller, J.C.; DeKelver, R.C.; Moehle, E.A.; Worden, S.E.; Mitchell, J.C.; Arnold, N.L.; Gopalan, S.; Meng, X.; et al. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 2009, 459, 437. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Liu, B.; Spalding, M.H.; Weeks, D.P.; Yang, B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 2012, 30, 390. [Google Scholar]
- Jung, Y.J.; Nogoy, F.M.; Lee, S.K.; Cho, Y.G.; Kang, K.K. Application of ZFN for site directed mutagenesis of rice SSIVa gene. Biotechnol. Bioprocess Eng. 2018, 23, 108–115. [Google Scholar] [CrossRef]
- Ran, Y.; Patron, N.; Kay, P.; Wong, D.; Buchanan, M.; Cao, Y.Y.; Sawbridge, T.; Davies, J.P.; Mason, J.; Webb, S.R.; et al. Zinc finger nuclease-mediated precision genome editing of an endogenous gene in hexaploid bread wheat (Triticum aestivum) using a DNA repair template. Plant Biotechn. J. 2018, 16, 2088–2101. [Google Scholar]
- Boch, J.; Scholze, H.; Schornack, S.; Landgraf, A.; Hahn, S.; Kay, S.; Lahaye, T.; Nickstadt, A.; Bonas, U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science 2009, 326, 1509–1512. [Google Scholar] [CrossRef]
- Doyle, E.L.; Booher, N.J.; Standage, D.S.; Voytas, D.F.; Brendel, V.P.; VanDyk, J.K.; Bogdanove, A.J. TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: Tools for TAL effector design and target prediction. Nucleic Acids Res. 2012, 40, W117–W122. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Huang, S.; Zhao, X.; Wright, D.A.; Carpenter, S.; Spalding, M.H.; Weeks, D.P.; Yang, B. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011, 39, 6315–6325. [Google Scholar]
- Ran, Y.; Liang, Z.; Gao, C. Current and future editing reagent delivery systems for plant genome editing. Sci. China Life Sci. 2017, 60, 490–505. [Google Scholar] [CrossRef]
- Zhang, Z.; Mao, Y.; Ha, S.; Liu, W.; Botella, J.R.; Zhu, J.K. A multiplex CRISPR/Cas9 platform for fast and efficient editing of multiple genes in Arabidopsis. Plant Cell Rep. 2016, 35, 1519–1533. [Google Scholar]
- Haun, W.; Coffman, A.; Clasen, B.M.; Demorest, Z.L.; Lowy, A.; Ray, E.; Retterath, A.; Stoddard, T.; Juillerat, A.; Cedrone, F.; et al. Improved soybean oil quality by targeted mutagenesis of the fatty acid desaturase 2 gene family. Plant Biotechnol. J. 2014, 12, 934–940. [Google Scholar] [CrossRef]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jansen, R.; Embden, J.D.V.; Gaastra, W.; Schouls, L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microboil. 2002, 43, 1565–1575. [Google Scholar]
- Shmakov, S.; Smargon, A.; Scott, D.; Cox, D.; Pyzocha, N.; Yan, W.; Abudayyeh, O.O.; Gootenberg, J.S.; Makarova, K.S.; Wolf, Y.I.; et al. Diversity and evolution of class 2 CRISPR–Cas systems. Nat. Rev. Microbiol. 2017, 15, 169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015, 13, 722. [Google Scholar] [CrossRef] [Green Version]
- Kleinstiver, B.P.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Joung, J.K. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529, 490. [Google Scholar] [CrossRef] [Green Version]
- Jackson, S.A.; McKenzie, R.E.; Fagerlund, R.D.; Kieper, S.N.; Fineran, P.C.; Brouns, S.J. CRISPR-Cas: Adapting to change. Science 2017, 356, eaal5056. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Alptekin, B.; Budak, H. CRISPR/Cas9 genome editing in wheat. Funct. Integr. Genom. 2018, 18, 31–41. [Google Scholar]
- Zhang, H.; Zhang, J.; Wei, P.; Zhang, B.; Gou, F.; Feng, Z.; Mao, Y.; Yang, L.; Zhang, H.; Xu, N.; et al. The CRISPR/C as9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 2014, 12, 797–807. [Google Scholar] [CrossRef]
- Shimatani, Z.; Kashojiya, S.; Takayama, M.; Terada, R.; Arazoe, T.; Ishii, H. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 441–443. [Google Scholar] [CrossRef]
- Zong, Y.; Wang, Y.; Li, C.; Zhang, R.; Chen, K.; Ran, Y. Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 438–440. [Google Scholar] [CrossRef]
- Li, J.; Sun, Y.; Du, J.; Zhao, Y.; Xia, L. Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol. Plant 2017, 10, 526–529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Z.; Zhang, K.; Chen, K.; Gao, C. Targeted mutagenesis in Zea mays using TALENs and the CRISPR/Cas system. J. Genet. Genom. 2014, 41, 63–68. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhu, L.; Zhao, B.; Zhao, Y.; Xie, Y.; Zheng, Z.; Li, Y.; Sun, J.; Wang, H. Development of a haploid-inducer mediated genome editing system for accelerating maize breeding. Mol. Plant 2019, 12, 597–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Gao, C. Recent advances in DNA-free editing and precise base editing in plants. Emerg. Top. Life Sci. 2017, 1, 61–168. [Google Scholar]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013, 41, e188. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.F.; Li, H.; Qin, R.Y.; Li, J.; Qiu, C.H.; Yang, Y.C.; Ma, H.; Li, L.; Wei, P.C.; Yang, J.B. Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci. Rep. 2015, 5, 11491. [Google Scholar] [CrossRef] [Green Version]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Miao, C.; Xiao, L.; Hua, K.; Zou, C.; Zhao, Y.; Bressan, R.A.; Zhu, J.K. Mutations in a subfamily of abscisic acid receptor genes promote rice growth and productivity. Proc. Natl. Acad. Sci. USA 2018, 115, 6058–6063. [Google Scholar] [CrossRef] [Green Version]
- Xie, E.; Li, Y.; Tang, D.; Lv, Y.; Shen, Y.; Cheng, Z. A strategy for generating rice apomixis by gene editing. J. Integr. Plant Biol. 2019. [Google Scholar] [CrossRef] [Green Version]
- Khanday, I.; Skinner, D.; Yang, B.; Mercier, R.; Sundaresan, V. A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 2019, 565, 91. [Google Scholar] [CrossRef]
- Pathak, B.; Zhao, S.; Manoharan, M.; Srivastava, V. Dual-targeting by CRISPR/Cas9 leads to efficient point mutagenesis but only rare targeted deletions in the rice genome. Biotech 2019, 9, 158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oliva, R.; Ji, C.; Atienza-Grande, G.; Huguet-Tapia, J.C.; Perez-Quintero, A.; Li, T.; Eom, J.S.; Li, C.; Nguyen, H.; Liu, B.; et al. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 2019, 37, 1344–1350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, S.; Jiang, J.; Liu, Y.; Meng, J.; Xu, S.; Tan, Y.; Li, Y.; Shu, Q.; Huang, J. Characterization and evaluation of OsLCT1 and OsNramp5 mutants generated through CRISPR/Cas9-mediated mutagenesis for breeding low Cd rice. Rice Sci. 2019, 26, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686. [Google Scholar] [CrossRef] [PubMed]
- Cermak, T.; Baltes, N.J.; Cegan, R.; Zhang, Y.; Voytas, D.F. High-frequency, precise modification of the tomato genome. Genome Biol. 2015, 16, 232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Liu, Z.-B.; Xing, A.; Moon, B.P.; Koellhoffer, J.P.; Huang, L.; Ward, R.T.; Clifton, E.; Falco, S.C.; Cigan, A.M. Cas9-guide RNA directed genome editing in soybean. Plant Physiol. 2015, 169, 960–970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Svitashev, S.; Schwartz, C.; Lenderts, B.; Young, J.K.; Mark Cigan, A. Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat. Commun. 2016, 7, 13274. [Google Scholar] [PubMed]
- Gil-Humanes, J.; Wang, Y.; Liang, Z.; Shan, Q.; Ozuna, C.V.; Sánchez-León, S.; Baltes, N.J.; Starker, C.; Barro, F.; Gao, C.; et al. High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 2017, 89, 1251–1262. [Google Scholar] [CrossRef] [Green Version]
- Tycko, J.; Myer, V.E.; Hsu, P.D. Methods for optimizing CRISPR-Cas9 genome editing specificity. Mol. Cell 2016, 63, 355–370. [Google Scholar] [PubMed] [Green Version]
- Xie, K.; Zhang, J.; Yang, Y. Genome-wide prediction of highly specific guide RNA spacers for CRISPR-Cas9-mediated genome editing in model plants and major crops. Mol. Plant 2014, 7, 923–926. [Google Scholar] [PubMed] [Green Version]
- Ranganathan, V.; Wahlin, K.; Maruotti, J.; Zack, D.J. Expansion of the CRISPRCas9 genome targeting space through the use of H1 promoter-expressed guide RNAs. Nat. Commun. 2014, 5, 4516. [Google Scholar] [PubMed] [Green Version]
- Wang, Z.P.; Xing, H.L.; Dong, L.; Zhang, H.Y.; Han, C.Y.; Wang, X.C.; Chen, Q.J. Egg cell-specific promoter-controlled CRISPR/Cas9 efficiently generates homozygous mutants for multiple target genes in Arabidopsis in a single generation. Genome Biol. 2015, 16, 144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Peng, Z.; Long, J.; Sosso, D.; Liu, B.; Eom, J.S.; Huang, S.; Liu, S.; Vera Cruz, C.; Frommer, W.B.; et al. Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 2015, 82, 632–643. [Google Scholar] [PubMed]
- Peng, A.; Chen, S.; Lei, T.; Xu, L.; He, Y.; Wu, L.; Yao, L.; Zou, X. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene Cs LOB 1 promoter in citrus. Plant Biotechnol. J. 2017, 15, 1509–1519. [Google Scholar] [CrossRef] [Green Version]
- Gao, J.; Wang, G.; Ma, S.; Xie, X.; Wu, X.; Zhang, X.; Wu, Y.; Zhao, P.; Xia, Q. CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol. Biol. 2015, 87, 99–110. [Google Scholar] [CrossRef]
- Han, X.; Liu, Z.; Jo, M.C.; Zhang, K.; Li, Y.; Zeng, Z.; Li, N.; Zu, Y.; Qin, L. CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation. Sci. Adv. 2015, 1, e1500454. [Google Scholar] [CrossRef] [Green Version]
- Ma, J.; Chen, J.; Wang, M.; Ren, Y.; Wang, S.; Lei, C.; Cheng, Z. Disruption of OsSEC3A increases the content of salicylic acid and induces plant defense responses in rice. J. Exp. Bot. 2017, 69, 1051–1064. [Google Scholar] [CrossRef] [Green Version]
- Mundt, C.C. Pyramiding for resistance durability: Theory and Practice. Phytopathology 2018, 108, 792–802. [Google Scholar]
- Zhou, H.; Liu, B.; Weeks, D.P.; Spalding, M.H.; Yang, B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014, 42, 10903–10914. [Google Scholar] [CrossRef]
- Dafny-Yelin, M.; Tzfira, T. Delivery of multiple transgenes to plant cells. Plant Physiol. 2007, 145, 1118–1128. [Google Scholar] [CrossRef] [Green Version]
- Que, Q.; Chilton, M.D.; de Fontes, C.M.; He, C.; Nuccio, M.; Zhu, T.; Wu, Y.; Chen, J.S.; Shi, L. Trait stacking in transgenic crops: Challenges and opportunities. GM Crops 2010, 1, 220–229. [Google Scholar] [PubMed]
- Zhu, S.; Li, Y.; Vossen, J.H.; Visser, R.G.F.; Jacobsen, E. Functional stacking of three resistance genes against Phytophthora infestans in potato. Transgenic Res. 2012, 21, 89–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voytas, D. Optimizing gene targeting in plants. Vitr. Cell. Dev. Biol. Anim. 2017, 53, S23. [Google Scholar]
- Zaman, Q.U.; Li, C.; Cheng, H.; Hu, Q. Genome editing opens a new era of genetic improvement in polyploid crops. Crop J. 2019, 7, 141–150. [Google Scholar] [CrossRef]
- Ali, Z.; Abulfaraj, A.; Idris, A.; Ali, S.; Tashkandi, M.; Mahfouz, M.M. CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 2015, 16, 238. [Google Scholar] [CrossRef] [Green Version]
- Zhan, M.; Coaker, G. Harnessing effector-triggered immunity fordurable disease resistance. Phytopathology 2017, 107, 912–919. [Google Scholar] [CrossRef] [Green Version]
- Zhang, R.; Liu, J.; Chai, Z.; Chen, S.; Bai, Y.; Zong, Y.; Chen, K.; Li, J.; Jiang, L.; Gao, C. Generation of herbicide tolerance traits and a new selectable marker in wheat using base editing. Nat. Plants 2019, 5, 480. [Google Scholar] [CrossRef]
- Huang, L.; Zhang, R.; Huang, G.; Li, Y.; Melaku, G.; Zhang, S.; Chen, H.; Zhao, Y.; Zhang, J.; Zhang, Y.; et al. Developing superior alleles of yield genes in rice by artificial mutagenesis using the CRISPR/Cas9 system. Crop J. 2018, 6, 475–481. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Kumar, J.; Alok, A.; Tuli, R. RNA-guided genome editing for target gene mutations in wheat. Genes Genomes Genet. 2013, 3, 2233–2238. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Pan, Q.; He, F.; Akhunova, A.; Chao, S.; Trick, H.; Akhunov, E. Transgenerational CRISPR-Cas9 activity facilitates multiplex gene editing in allopolyploid wheat. CRISPR J. 2018, 1, 65–74. [Google Scholar]
- Liao, S.; Qin, X.; Luo, L.; Han, Y.; Wang, X.; Usman, B.; Nawaz, G.; Zhao, N.; Liu, Y.; Li, R. CRISPR/Cas9-Induced Mutagenesis of Semi-Rolled Leaf 1, 2 Confers Curled Leaf Phenotype and Drought Tolerance by Influencing Protein Expression Patterns and ROS Scavenging in Rice (Oryza sativa L.). Agronomy 2019, 9, 728. [Google Scholar] [CrossRef] [Green Version]
- Qiao, F.; Yang, Q.; Wang, C.L.; Fan, Y.L.; Wu, X.F.; Zhao, K.J. Modification of plant height via RNAi suppression of OsGA20ox2 gene in rice. Euphytica 2007, 158, 35–45. [Google Scholar] [CrossRef]
- Gothandam, K.M.; Nalini, E.; Karthikeyan, S.; Shin, J.S. OsPRP3, a flower specific proline-rich protein of rice, determines extracellular matrix structure of floral organs and its overexpression confers cold-tolerance. Plant Mol. Biol. 2010, 72, 125. [Google Scholar] [CrossRef] [PubMed]
- Khandagale, K.S.; Zanan, R.L.; Mathure, S.V.; Nadaf, A.B. Haplotype variation of Badh2 gene, unearthing of a new fragrance allele and marker development for non-basmati fragrant rice ‘Velchi’ (Oryza sativa L.). Agri Gene 2017, 6, 40–46. [Google Scholar] [CrossRef]
- Sternberg, S.H.; Haurwitz, R.E.; Doudna, J.A. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. RNA 2012, 18, 661–672. [Google Scholar] [CrossRef] [Green Version]
- Čermák, T.; Curtin, S.J.; Gil-Humanes, J.; Čegan, R.; Kono, T.J.; Konečná, E.; Belanto, J.J.; Starker, C.G.; Mathre, J.W.; Greenstein, R.L.; et al. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 2017, 29, 1196–1217. [Google Scholar] [CrossRef] [Green Version]
- Qi, W.; Zhu, T.; Tian, Z.; Li, C.; Zhang, W.; Song, R. High-efficiency CRISPR/Cas9 multiplex gene editing using the glycine tRNA-processing system-based strategy in maize. BMC Biotechnol. 2016, 16, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Q.; Xu, K.; Xing, J.; Zhang, T.; Wang, X.; Wei, Z.; Ren, C.; Liu, Z.; Shao, S.; Zhang, Z. Multiplex CRISPR/Cas9-based genome engineering enhanced by Drosha-mediated sgRNA-shRNA structure. Sci. Rep. 2016, 6, 38970. [Google Scholar] [CrossRef] [Green Version]
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalan, C.; Cho, S.W.; Kim, H.; Kim, S.G.; Kim, S.T.; Choe, S.; Kim, J.S. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 2015, 33, 1162–1164. [Google Scholar] [CrossRef]
- Guilinger, J.P.; Thompson, D.B.; Liu, D.R. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014, 32, 577. [Google Scholar] [CrossRef]
- Battraw, M.J.; Hall, T.C. Histochemical analysis of CaMV 35S promoter-β-glucuronidase gene expression in transgenic rice plants. Plant Mol. Biol. 1990, 15, 527–538. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Hong, W.; Wu, J.; Wang, Y.; Ji, S.; Zhu, S.; Wei, C.; Zhang, J.; Li, Y. A viral protein promotes host SAMS1 activity and ethylene production for the benefit of virus infection. Elife 2017, 6, e27529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, S.H.; Yi, N.; Kim, Y.S.; Jeong, M.H.; Bang, S.W.; Choi, Y.D.; Kim, J.K. Analysis of five novel putative constitutive gene promoters in transgenic rice plants. J. Exp. Bot. 2010, 61, 2459–2467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, Y.; Zhang, Z.; Feng, Z.; Wei, P.; Zhang, H.; Botella, J.R.; Zhu, J.K. Development of germ-line-specific CRISPR-Cas9 systems to improve the production of heritable gene modifications in Arabidopsis. Plant Biotechnol. J. 2016, 14, 519–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mikami, M.; Toki, S.; Endo, M. Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol. Biol. 2015, 88, 561–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borghi, L. Inducible gene expression systems for plants. In Plant Developmental Biology; Humana Press: Totowa, NJ, USA, 2010; pp. 65–75. [Google Scholar]
- Decaestecker, W.; Buono, R.A.; Pfeiffer, M.L.; Vangheluwe, N.; Jourquin, J.; Karimi, M.; Van Isterdael, G.; Beeckman, T.; Nowack, M.K.; Jacobs, T.B. CRISPR-TSKO facilitates efficient cell type-, tissue-, or organ-specific mutagenesis in Arabidopsis. BioRxiv 2018, 474981. [Google Scholar] [CrossRef]
- Ren, Q.; Zhong, Z.; Wang, Y.; You, Q.; Li, Q.; Yuan, M.; He, Y.; Qi, C.; Tang, X.; Zheng, X.; et al. Bidirectional promoter based CRISPR-Cas9 systems for plant genome editing. Front. Plant Sci. 2019, 10, 1173. [Google Scholar] [CrossRef]
- Xie, K.; Minkenberg, B.; Yang, Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. USA 2015, 112, 3570–3575. [Google Scholar] [CrossRef] [Green Version]
- Schinkel, H.; Schillberg, S. Genome editing: Intellectual property and product development in plant biotechnology. Plant Cell Rep. 2016, 35, 1487–1491. [Google Scholar] [CrossRef]
- Elison, G.L.; Xue, Y.; Song, R.; Acar, M. Insights into bidirectional gene expression control using the canonical GAL1/GAL10 promoter. Cell Rep. 2018, 25, 737–748. [Google Scholar] [CrossRef] [Green Version]
- Peer, R.; Rivlin, G.; Golobovitch, S.; Lapidot, M.; Gal-On, A.; Vainstein, A.; Tzfira, T.; Flaishman, M.A. Targeted mutagenesis using zinc-finger nucleases in perennial fruit trees. Planta 2015, 241, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Mahfouz, M.M.; Li, L.; Shamimuzzaman, M.; Wibowo, A.; Fang, X.; Zhu, J.K. De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc. Natl. Acad. Sci. USA 2011, 108, 2623–2628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curtin, S.J.; Zhang, F.; Sander, J.D.; Haun, W.J.; Starker, C.; Baltes, N.J.; Reyon, D.; Dahlborg, E.J.; Goodwin, M.J.; Coffman, A.P.; et al. Targeted mutagenesis of duplicated genes in soybean with zinc-finger nucleases. Plant Physiol. 2011, 156, 466–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, H.; Zeng, X.; Zhao, M.; Cui, X.; Wang, Q.; Yang, H.; Cheng, H.; Yu, D. Efficient targeted mutagenesis in soybean by TALENs and CRISPR/Cas9. J. Biotechnol. 2016, 217, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A• T to G• C in genomic DNA without DNA cleavage. Nature 2017, 551, 464. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, C.; Liu, W.; Gao, W.; Liu, C.; Song, G.; Li, W.X.; Mao, L.; Chen, B.; Xu, Y.; et al. An alternative strategy for targeted gene replacement in plants using a dual-sgRNA/Cas9 design. Sci. Rep. 2016, 6, 23890. [Google Scholar] [CrossRef]
- Sera, T. Zinc-finger-based artificial transcription factors and their applications. Adv. Drug Deliv. Rev. 2009, 61, 513–526. [Google Scholar] [CrossRef]
- Perez-Pinera, P.; Ousterout, D.; Brunger, J.; Farin, A.; Glass, K.; Guilak, F.; Crawford, G.; Hartemink, A.; Gersback, C. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nat. Methods 2013, 10, 239–242. [Google Scholar] [CrossRef]
- Li, G.; Jain, R.; Chern, M.; Pham, N.T.; Martin, J.A.; Wei, T.; Schackwitz, W.S.; Lipzen, A.M.; Duong, P.Q.; Jones, K.C.; et al. The sequences of 1504 mutants in the model rice variety kitaake facilitate rapid functional genomics studies. The Plant Cell 2017, 29, 1218–1231. [Google Scholar] [CrossRef] [Green Version]
- Osborn, T.C.; Pires, J.C.; Birchler, J.A.; Auger, D.L.; Chen, Z.J.; Lee, H.S.; Comai, L.; Madlung, A.; Doerge, R.W.; Colot, V.; et al. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 2003, 19, 141–147. [Google Scholar] [CrossRef]
- Braatz, J.; Harloff, H.J.; Mascher, M.; Stein, N.; Himmelbach, A.; Jung, C. CRISPR-Cas9 targeted mutagenesis leads to simultaneous modification of different homoeologous gene copies in polyploid oilseed rape (Brassica napus). Plant Physiol. 2017, 174, 935–942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunet, E.; Simsek, D.; Tomishima, M.; DeKelver, R.; Choi, V.M.; Gregory, P.; Urnov, F.; Weinstock, D.M.; Jasin, M. Chromosomal translocations induced at specified loci in human stem cells. Proc. Natl. Acad. Sci. USA 2009, 106, 10620–10625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.J.; Kweon, J.; Kim, E.; Kim, S.; Kim, J.S. Targeted chromosomal duplications and inversions in the human genome using zinc finger nucleases. Genome Res. 2012, 22, 539–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kweon, J.; Kim, A.; Chon, J.K.; Yoo, J.Y.; Kim, H.J.; Kim, S.; Lee, C.; Jeong, E.; Chung, E. A library of TAL effector nucleases spanning the human genome. Nat. Biotechnol. 2013, 31, 251–258. [Google Scholar]
- Sander, J.D.; Joung, J.K. CRISPR-Cas systems for genome editing, regulation and targeting. Nat. Biotechnol. 2014, 32, 347. [Google Scholar] [CrossRef]
- Zhang, X.H.; Tee, L.Y.; Wang, X.G.; Huang, Q.S.; Yang, S.H. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol. Ther.-Nucleic Acids 2015, 4, 264. [Google Scholar] [CrossRef]
- Tsai, S.Q.; Zheng, Z.; Nguyen, N.T.; Liebers, M.; Topkar, V.V.; Thapar, V.; Aryee, M.J. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Bitechnol. 2015, 33, 187. [Google Scholar]
- Ren, X.; Yang, Z.; Xu, J.; Sun, J.; Mao, D.; Hu, Y.; Deng, P. Enhanced specificity and efficiency of the CRISPR/Cas9 system with optimized sgRNA parameters in Drosophila. Cell Rep. 2014, 9, 1151–1162. [Google Scholar] [CrossRef] [Green Version]
- Vats, S.; Kumawat, S.; Kumar, V.; Patil, G.B.; Joshi, T.; Sonah, H.; Sharma, T.R.; Deshmukh, R. Genome Editing in Plants: Exploration of Technological Advancements and Challenges. Cells 2019, 8, 1386. [Google Scholar]
- Feng, Z.; Mao, Y.; Xu, N.; Zhang, B.; Wei, P.; Yang, D.L.; Wang, Z.; Zhang, Z.; Zheng, R.; Yang, L.; et al. Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 4632–4637. [Google Scholar] [CrossRef] [Green Version]
- Maaß, O.; Consmüller, N.; Kehlenbeck, H. Socioeconomic Impact of Genome Editing on Agricultural Value Chains: The Case of Fungal-Resistant and Coeliac-Safe Wheat. Sustainability 2019, 11, 6421. [Google Scholar]
- Agapito-Tenfen, S.Z.; Okoli, A.S.; Bernstein, M.J.; Wikmark, O.G.; Myhr, A.I. Revisiting risk governance of GM plants: The need to consider new and emerging gene-editing techniques. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Bortesi, L.; Zhu, C.; Zischewski, J.; Perez, L.; Bassié, L.; Nadi, R.; Forni, G.; Lade, S.B.; Soto, E.; Jin, X.; et al. Patterns of CRISPR/Cas9 activity in plants, animals and microbes. Plant Biotechnol. J. 2016, 14, 2203–2216. [Google Scholar] [CrossRef] [PubMed]
- Wolt, J.D.; Wang, K.; Yang, B. The regulatory status of genome-edited crops. Plant Biotechnol. J. 2016, 14, 510–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davison, J.; Ammann, K. New GMO regulations for old: Determining a new future for EU crop biotechnology. GM Crops Food 2017, 8, 13–34. [Google Scholar]
- Globus, R.; Qimrom, U. A technological and regulatory outlook on CRISPR crop editing. J. Cell. Biochem. 2018, 119, 1291–1298. [Google Scholar] [CrossRef]

| Plant Species | Delivery Mode | Target Gene(s) | Gene Function | Vector Used | Promoter Used | Reference |
|---|---|---|---|---|---|---|
| Maize | Agrobacterium-transformation | ALS2 | A key enzyme for the biosynthesis of branched-chain amino acids (major targets for herbicides) | UBI:Cas9 T-DNA vector | ZmU1 | [9] |
| Maize | Agrobacterium-mediated transformation | PSY1 | Phytoene synthase | pMD18-T | ZmU6 | [10] |
| Maize | Protoplast transformation | Zmzb7 | Encodes IspH protein for methyl-D-erythritol-4- phosphate (MEP) Pathway | pEasy-Blunt simple vector | ZmU3 | [11] |
| Maize | Agrobacterium-mediated transformation | ARGOS8 | Increased grain yield under drought stress | sgRNA-Cas9 | ZmU6 | [12] |
| Wheat | Agrobacterium-mediated transformation | TaMLO | Mildew resistance locus | pUC-T vector (CWBIO) | TaU6 | [13] |
| Wheat | Biolistic transformation | TaMLO-A1 | Powdery mildew resistance negative regulator | pJIT163 | TaU6 | [14] |
| Wheat | Agrobacterium-mediated transformation | TaVIT2 | Fe content | p416-MET25 | HMW-GLU | [15] |
| Wheat | Biolistic transformation | TaEDR1 | Disease resistance Against powdery mildew | pJIT163-Ubi-Cas9 | TaU6 | [16] |
| Wheat | Biolistic bombardment | TaGW2 | Grain weight negative Regulator | pET28a-Cas9-His | TaU6 | [17] |
| Wheat | Biolistic transformation | Alpha-gliadin | Low-gluten | pANIC-6E | TaU6 | [18] |
| Rice | Agrobacterium-mediated transformation | osPPKL1 | increases length and yield | pCAMBIA1300S | CaMV 35S | [19] |
| Rice | Agrobacterium-mediated transformation | OsMPK5 | Various abiotic stress tolerance and disease resistance | pRGE3 and pRGE6 | OsU3 and OsU6 | [20] |
| Rice | Agrobacterium-mediated transformation | TMS5 | Negative regulator of thermo-sensitive genic male sterility | TMS5as | OsU3 | [21] |
| Rice | Agrobacterium-mediated transformation | ALS | A key enzyme for the biosynthesis of branched-chain amino acids, (major targets for herbicides) | pCXUN-Cas9-gRNA1-gRNA2-armed donor vector | OsU3 | [22] |
| Rice | Biolistic transformation | EPSPS | A key enzyme of aromatic amino acids biosynthesis | pHUN411 | OsU3 | [23] |
| Rice | Agrobacterium-mediated transformation | CSA | Negative regulator of photoperiod-sensitive genic male sterility | CH-CRISPR/Cas9-CSA, Gateway-CRISPR/Cas9-CSA | OsU6a | [24] |
| Rice | Agrobacterium-mediated transformation | DEP1 | Regulators of inflorescence Architecture of plant height | pYLCRISPR/Cas9(I) | OsU6a | [25] |
| Rice | Electroporation | ERF922 | Rice blast resistance negative regulator | C-ERF922 | OsU6a | [26] |
| Rice | Agrobacterium-mediated transformation | NRT1.1B | Nitrogen transporter | PCSGAPO1 | OsU6 | [27] |
| Rice | Agrobacterium-mediated transformation | OsHAK-1 | Low cesium accumulation | pH-Ubi-Cas9-7 | OsUbi | [28] |
| Rice | Electroporation | SBEIIb | high-amylose | pCXUNCas9 | OsU3 | [29] |
| Rice | Agrobacterium-mediated transformation | OsNramp5 | Low Cd-accumulation | pYLCRISPR/Cas9Pubi-H | OsU3, OsU6 | [30] |
| Rice | Agrobacterium-mediated transformation | eIF4G | Resistance to rice tungrospherical virus | pCas9-eIF4G-gRNA | TaU6 | [31] |
| Rice | Agrobacterium-mediated transformation | OsPRX2 | Potassium deficiency tolerance | pCAMBIA1301 | OsPRX2 | [32] |
| Rice | Agrobacterium-mediated transformation | Waxy | Amylose content | CRISPR/Cas9 vector | OsU6 | [33] |
| Rice | Agrobacterium-mediated transformation | NGv1 | Reduction of off-target effects | APOBEC-UGI | OsU6 | [34] |
| Rice | Agrobacterium-mediated transformation | Tos17 | retrotransposon | CRISPR/Cas9 vectors | OsU6 | [35] |
| Rice | Agrobacterium-mediated transformation | ISA1 | isoamylase-type debranching enzyme | VK005 | OsU6 | [36] |
| Rice | Agrobacterium-mediated transformation | OsRR22 | salinity tolerance | pYLCRISPR/Cas9Pubi-H | OsU6 | [37] |
| Rice | Agrobacterium-mediated transformation | OsITPK6 | Low phytic acid | pHun4c12s | OsU6 | [38] |
| Barley | Agrobacterium-mediated transformation | HvPM19 | ABA-inducible plasma membrane protein | pAGM4723 | TaU6 | [39] |
| Plant Species | Delivery Mode | Target Gene(s) | Gene Function | Vector Used | Promoter Used | Reference |
|---|---|---|---|---|---|---|
| Maize | Agrobacterium- transformation | ZmIPK1A, ZmIPK and ZmMRP4 | Phytic acid synthesis | pEasy Blunt vector | ZmU3 | [62] |
| Maize | Biolistic-mediated transformation | LIG, MS26, MS26, MS45, LIG, MS26, MS45 | LIG (liguleless) MS26 and 45 (male sterility) | Cas9 DNA vector | ZmU6 | [9] |
| Maize | Agrobacterium-mediated transformation | ZmLG1, UB2, and UB3 | Development of a haploid-inducer mediated genome editing system | pCPB | CaMV 35S | [63] |
| Wheat | PEG4000-mediated transformation | TaDEP1, TaGASR7, TaLOX2, TaNAC2, TaPIN1, TaGW2 | Inflorescence architecture and plant height regulator, lipoxygenase, grain weight negative regulator | pJIT163 | TaU6 | [64] |
| Wheat | Agrobacterium-mediated transformation | TaDREB2 and TaERF3 | Drought resistance | pJIT163-2NLSCas9 | TaU6 | [57] |
| Rice | Agrobacterium-mediated transformation | OsSWEET11, OsSWEET14 | sucrose efflux transporter | pTOPO/D | OsU6 | [65] |
| Rice | Biolistic transformation | OsBADH2, Os02g23823, OsMPK2 | Responsible for aroma, a basic helix–loop–helix (bHLH) transcription factor, a mitogen-activated protein kinase | pJIT163 | OsU3 | [13] |
| Rice | Biolistic transformation | OsMPK2, OsDEP1 | Yield under stress | pJIT163 | OsU3 | [13] |
| Rice | Agrobacterium-mediated transformation | OsDERF1, OsPMS3, OsEPSPS, OsMSH1, OsMYB5 | Drought tolerance | sgRNA-Cas9 | OsU3 | [58] |
| Rice | Agrobacterium-mediated transformation | OsPDS, OsPMS3, OsEPSPS, OsDERF1, OsMSH1, OsMYB5, OsMYB1, OsROC5, OsSPP and OsYSA | (OsPDS) pigment synthesis, (OsEPSPS) synthesis of aromatic amino acid, (OsMSH1) DNA mismatch repair protein, (OsROC5) Rice Outermost Cell-specific gene5, (OsDERF1) AP2 domain containing protein, (OsYSA) pentatricopeptide repeat domain containing protein | WDV2-ACT1 and WDV2-GST | 35s | [25] |
| Rice | Agrobacterium-mediated transformation | OsAOX1a, OsAOX1b, OsAOX1c, OsBEL | Various abiotic stress tolerance | GATEWAY-based vector | OsU3 | [66] |
| Rice | Agrobacterium-mediated transformation | OsU3, OsU6a, OsU6b, OsU6c | OsWaxy; amylase synthase | pCAMBIA1300 | OsU3, OsU6b, and OsU6c | [67] |
| Rice | Agrobacterium-mediated transformation | GS3, GW2, GW5, TGW6 | Grain Size 3 (GS3), grain width 2 (GW2), grain width 5 (GW5) and thousand-grain weight 6 (TGW6), negatively regulated grain weight | pHUN412 | GW2-OsU3 GW5-OsU6 TGW6-TaU3 | [6] |
| Rice | Agrobacterium-mediated transformation | DEP1, Gn1a, IPA1, GS3 | (DEP1) inflorescence architecture and plant height, (Gn1a) grain number negative regulator, (IPA1) plant architecture regulator, (GS3) negative regulator of grain size | pYLCRISPR/Cas9(I) | OsU6a | [24] |
| Rice | Agrobacterium-mediated transformation | GW2GW5 TGW6 | Grain weight negative regulator | pHUN412 vector | OsU3, OsU6 and TaU3 | [6] |
| Rice | Agrobacterium-mediated transformation | PYL1–PYL6 and PYL12(gp-1), PYL7–PYL11 and PYL13(gp-2) | best growth and improved grain productivity | PCAMBIA1300 | OsU3, OsU6 | [68] |
| Rice | Agrobacterium-mediated transformation | SPO11-1, REC8, OSD1, MATL | Introduction of apomixis | pC1300-Cas9 | OsU6 | [69] |
| Rice | Agrobacterium-mediated transformation | BBM1, BBM2 and BBM3 | Redirection for asexual propagation through seeds | pCRISPR BBM | OsU6 | [70] |
| Rice | Transformation by gene gun | GUS, PDS, Chalk5 | Investigation of the efficiency of CRISPR/Cas9 in creating genomic deletions | pRGE32, pJU24, pJU34 and pJU46 | OsU3 | [71] |
| Rice | Agrobacterium-mediated transformation | SWEET11, SWEET13 and SWEET14 | resistance to bacterial blight | pBY02-ZmUbiP-OsCas9 | ZmUbi | [72] |
| Rice | Agrobacterium-mediated transformation | OsLCT1 and OsNramp5 | Low cadmium (Cd) | pHun4c12s | OsU6 | [73] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ansari, W.A.; Chandanshive, S.U.; Bhatt, V.; Nadaf, A.B.; Vats, S.; Katara, J.L.; Sonah, H.; Deshmukh, R. Genome Editing in Cereals: Approaches, Applications and Challenges. Int. J. Mol. Sci. 2020, 21, 4040. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21114040
Ansari WA, Chandanshive SU, Bhatt V, Nadaf AB, Vats S, Katara JL, Sonah H, Deshmukh R. Genome Editing in Cereals: Approaches, Applications and Challenges. International Journal of Molecular Sciences. 2020; 21(11):4040. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21114040
Chicago/Turabian StyleAnsari, Waquar A., Sonali U. Chandanshive, Vacha Bhatt, Altafhusain B. Nadaf, Sanskriti Vats, Jawahar L. Katara, Humira Sonah, and Rupesh Deshmukh. 2020. "Genome Editing in Cereals: Approaches, Applications and Challenges" International Journal of Molecular Sciences 21, no. 11: 4040. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21114040





