Regulation of Three Key Kinases of Brassinosteroid Signaling Pathway
Abstract
:1. Introduction
2. BRI1, the BR Receptor
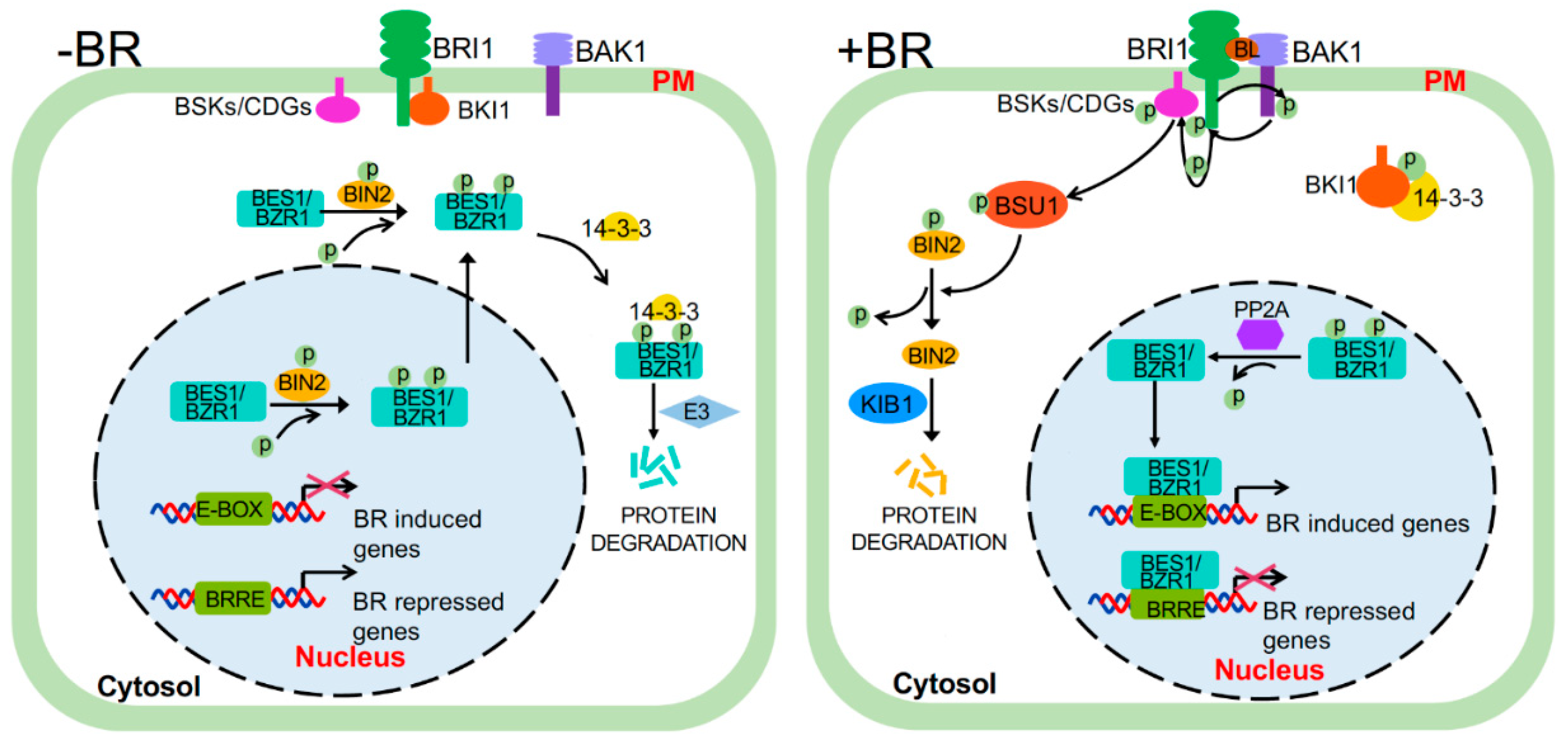
2.1. Maintaining the Inactive State in the Absence of BR
2.2. BRI1 Activation
2.3. Attenuation and Deactivation
2.4. Regulating the Abundance of BRI1 on the PM
2.4.1. Trafficking from the ER to the PM
2.4.2. Targeting to Unique PM Nano/Microdomains
2.4.3. Endocytosis
2.4.4. The Endocytic Pathway to the Vacuole for Degradation
2.4.5. Endocytic Recycling
3. BAK1, the Coreceptor
3.1. Phosphorylation of BAK1
3.2. Regulating BAK1 Availability for BRI1 Interaction
3.3. BAK1 Regulation by Other Mechanisms
4. BIN2, the Negative Regulator
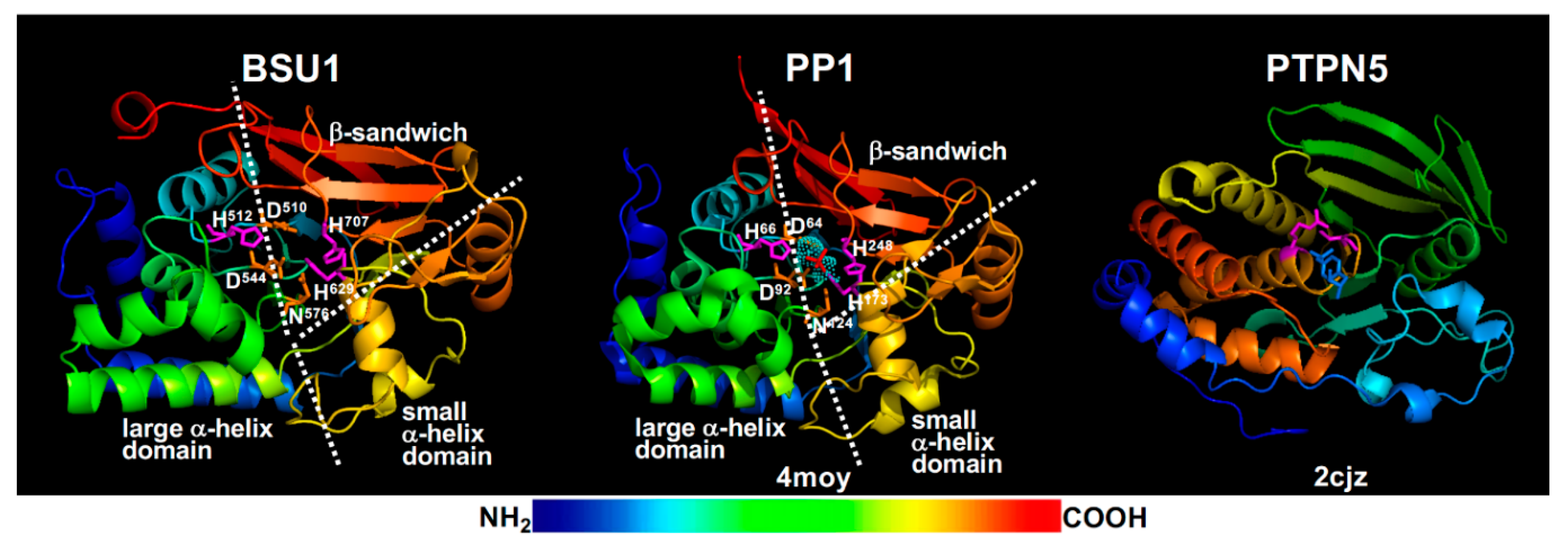
4.1. BIN2 Regulation by Dephosphorylation
4.2. BIN2 Regulation by Other Post-Translational Modifications
4.3. BIN2 Regulation by Protein–Protein Interactions
4.4. BIN2 Regulation by Subcellular Localization
4.5. BIN2 Inhibition by Degradation
5. Conclusions and Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AA | amino acids |
| ABA | abscisic acid |
| ABI1/2 | ABA-Insensitive1/2 |
| ALIX | apoptosis-linked gene 2-interacting protein X |
| AP2 | adapter protein 2 |
| ARF-GEF | ADP-ribosylation factor-guanine nucleotide exchange factor |
| AtFlot1 | Arabidopsis thaliana Flot1 |
| BAK1 | BRI1-Associated receptor Kinase1 |
| BARK1 | BAK1-Associated Receptor-like Kinase1 |
| BES1 | bri1-EMS suppressor1 |
| BIG3/5 | brefeldin A-inhibited guanine nucleotide-exchange protein3/5 |
| BIK1 | Botrytis-Induced Kinase1 |
| BIL4 | Brassinazole-Insensitive-Long hypocotyl4 |
| BIN2 | Brassinosteroid-Insensitive2 |
| BIR1-3 | BAK1-Interacting Receptor-like kinase1-3 |
| BKI1 | BRI1 Kinase Inhibitor1 |
| BLK1-3 | BARK1-Like Kinase1-3 |
| BR | Brassinosteroid |
| BRI1 | Brassinosteroid-Insensitive1 |
| BRL1-3 | BRI1-Like1-3 |
| BRRE | BR-response element |
| BSK1/3 | BR-Signaling Kinase1/3 |
| BSL1-3 | BSU1-Like1-3 |
| BSU1 | bri1 suppressor1 |
| BZR1 | Brassinazole-Resistant1 |
| CDG1 | Constitutive Differential Growth1 |
| CIE | clathrin-independent endocytosis |
| CLASP | cytoplasmic linker-associated protein |
| CME | clathrin-mediated endocytosis |
| COP1 | Constitutive Photomorphogenesis1 |
| CRY1 | Cryptochrome1 |
| CT | C-terminal tail |
| det2/DET3 | de-etiolated2/3 |
| DW1 | Dwarfing1 |
| EFR | Elongation Factor-Tu Receptor |
| elg1-D | elongated 1-Dominant |
| ER | endoplasmic reticulum |
| ERAD | ER-associated degradation |
| ERQC | ER-quality control |
| ESCRT | endosomal sorting complex required for transport |
| EXO70A1 | exocyst subunit 70A1 |
| FLS2 | Flagellin Sensing2 |
| G-loop | glycine-rich loop |
| AtGRXC2 | Arabidopsis thaliana glutaredoxin C2 |
| GSK3 | Glycogen Synthase Kinase3 |
| GW5 | grain width and weight5 |
| HDA6 | histone deacetylase6 |
| HSP90 | heat shock protein 90 |
| HsPTPN5 | human PTP non-receptor type 5 |
| HY5 | long hypocotyl5 |
| ID | island domain |
| ILVs | intraluminal vesicles |
| JM | juxtamembrane |
| KIB1 | Kink suppressed in bzr1-1D1 |
| LE/MVBs | late endosomes/multivesicular bodies |
| LRR-RLK | leucine-rich repeat receptor-like kinase |
| MS | mass spectrometry |
| MSBP1 | membrane steroid binding protein1 |
| NES | nuclear export signal |
| NLS | nuclear localization signal |
| NO | nitric oxide |
| OPS | OCTOPUS |
| OsGSK3 | Oryza sativa GSK3-like kinase3 |
| OsREM4.1 | Oryza sativa Remorin 4.1 |
| PIF3 | Phytochrome-Interacting Factor3 |
| PM | plasma membrane |
| POLAR | polar localization during asymmetric division and redistribution |
| PP2A | protein phosphatase 2A |
| PPKL | protein phosphatase with Kelch-like domains |
| PSP | protein Ser/Thr phosphatase |
| PTP/DSPP | protein tyrosine phosphatase/dual specificity protein phosphatase |
| pTyr | phosphorylated tyrosine residue |
| PUB12/13 | plant U-box protein12/13 |
| QC | quality control |
| qGL3 | quantitative trait locus regulating grain length3 |
| ROS | reactive oxygen species |
| R-spine | regulatory-spine |
| SbBIN2 | Sorghum bicolor BIN2 homolog |
| SBI1 | suppressor of bri1 1 |
| SERK | Somatic Embryogenesis Receptor Kinase |
| Sirt1/7 | Sirtuin1/7 |
| SNARE | soluble N-ethylmaleimide-sensitive factor attachment proteins receptor |
| SNX1 | sorting nexin1 |
| sobir7-1 | suppressor of bir1 7-1 |
| SPA | Suppressor of phyA-105 |
| TGN/EE | trans-Golgi network/early endosome |
| TPC | TPLATE complex |
| TTL | Tetratricopeptide Thioredoxin-Like |
| TWD1 | Twisted Dwarf1 |
| ucu | ultracurvata |
| V-ATPase | Vacuolar ATPase |
| VH1 | Vascular Highway1 |
References
- Clouse, S.D.; Sasse, J.M. Brassinosteroids: Essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 427–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.Y.; Bai, M.Y.; Oh, E.; Zhu, J.Y. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu. Rev. Genet. 2012, 46, 701–724. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Li, L.; Aluru, M.; Aluru, S.; Yin, Y. Mechanisms and networks for brassinosteroid regulated gene expression. Curr. Opin. Plant Biol. 2013, 16, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.J.; Zhang, C.; Lu, Y.N.; Jin, J.Q.; Wang, X.L. The mechanisms of brassinosteroids’ action: From signal transduction to plant development. Mol. Plant 2011, 4, 588–600. [Google Scholar] [CrossRef] [Green Version]
- Nawaz, F.; Naeem, M.; Zulfiqar, B.; Akram, A.; Ashraf, M.Y.; Raheel, M.; Shabbir, R.N.; Hussain, R.A.; Anwar, I.; Aurangzaib, M. Understanding brassinosteroid-regulated mechanisms to improve stress tolerance in plants: A critical review. Environ. Sci. Pollut. Res. Int. 2017, 24, 15959–15975. [Google Scholar] [CrossRef]
- Saini, S.; Sharma, I.; Pati, P.K. Versatile roles of brassinosteroid in plants in the context of its homoeostasis, signaling and crosstalks. Front Plant Sci. 2015, 6, 950. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.-F.; Lu, J.; Yu, J.-W.; Zhang, C.-Q.; He, J.-X.; Liu, Q.-Q. The brassinosteroid-regulated transcription factors BZR1/BES1 function as a coordinator in multisignal-regulated plant growth. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 561–571. [Google Scholar] [CrossRef]
- Li, J.; Nagpal, P.; Vitart, V.; McMorris, T.C.; Chory, J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science 1996, 272, 398–401. [Google Scholar] [CrossRef]
- Li, J.; Chory, J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell 1997, 90, 929–938. [Google Scholar] [CrossRef] [Green Version]
- Nolan, T.M.; Vukasinovic, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multidimensional regulators of plant growth, development, and stress responses. Plant Cell 2020, 32, 295–318. [Google Scholar] [CrossRef] [Green Version]
- Planas-Riverola, A.; Gupta, A.; Betegon-Putze, I.; Bosch, N.; Ibanes, M.; Cano-Delgado, A.I. Brassinosteroid signaling in plant development and adaptation to stress. Development 2019, 146, dev151894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belkhadir, Y.; Jaillais, Y. The molecular circuitry of brassinosteroid signaling. New Phytol. 2015, 206, 522–540. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Seto, H.; Fujioka, S.; Yoshida, S.; Chory, J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 2001, 410, 380–383. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wen, J.; Lease, K.A.; Doke, J.T.; Tax, F.E.; Walker, J.C. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 2002, 110, 213–222. [Google Scholar] [CrossRef] [Green Version]
- Russinova, E.; Borst, J.-W.; Kwaaitaal, M.; Caño-Delgado, A.; Yin, Y.; Chory, J.; de Vries, S.C. Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1). Plant Cell 2004, 16, 3216–3229. [Google Scholar] [CrossRef] [Green Version]
- Santiago, J.; Henzler, C.; Hothorn, M. Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 2013, 341, 889–892. [Google Scholar] [CrossRef]
- Ma, X.; Xu, G.; He, P.; Shan, L. SERKing coreceptors for receptors. Trends. Plant Sci. 2016, 21, 1017–1033. [Google Scholar] [CrossRef]
- Nam, K.H.; Li, J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell 2002, 110, 203–212. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Han, Z.; Tang, J.; Hu, Z.; Chai, C.; Zhou, B.; Chai, J. Structure reveals that BAK1 as a co-receptor recognizes the BRI1-bound brassinolide. Cell Res. 2013, 23, 1326–1329. [Google Scholar] [CrossRef]
- Wang, X.; Chory, J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science 2006, 313, 1118–1122. [Google Scholar] [CrossRef]
- Wang, X.; Kota, U.; He, K.; Blackburn, K.; Li, J.; Goshe, M.B.; Huber, S.C.; Clouse, S.D. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev. Cell 2008, 15, 220–235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Goshe, M.B.; Soderblom, E.J.; Phinney, B.S.; Kuchar, J.A.; Li, J.; Asami, T.; Yoshida, S.; Huber, S.C.; Clouse, S.D. Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis Brassinosteroid-Insensitive1 receptor kinase. Plant Cell 2005, 17, 1685–1703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Li, X.; Meisenhelder, J.; Hunter, T.; Yoshida, S.; Asami, T.; Chory, J. Autoregulation and homodimerization are involved in the activation of the plant steroid receptor BRI1. Dev. Cell 2005, 8, 855–865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, W.; Kim, T.W.; Oses-Prieto, J.A.; Sun, Y.; Deng, Z.; Zhu, S.; Wang, R.; Burlingame, A.L.; Wang, Z.Y. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science 2008, 321, 557–560. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.W.; Guan, S.; Burlingame, A.L.; Wang, Z.Y. The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3-like kinase BIN2. Mol. Cell 2011, 43, 561–571. [Google Scholar] [CrossRef] [Green Version]
- Lin, W.; Lu, D.; Gao, X.; Jiang, S.; Ma, X.; Wang, Z.; Mengiste, T.; He, P.; Shan, L. Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc. Natl. Acad. Sci. USA 2013, 110, 12114–12119. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.W.; Guan, S.; Sun, Y.; Deng, Z.; Tang, W.; Shang, J.X.; Sun, Y.; Burlingame, A.L.; Wang, Z.Y. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 2009, 11, 1254–1260. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Li, Y.; Cao, D.M.; Yang, H.; Oh, E.; Bi, Y.; Zhu, S.; Wang, Z.Y. The F-box protein KIB1 mediates brassinosteroid-induced inactivation and degradation of GSK3-like kinases in Arabidopsis. Mol. Cell 2017, 66, 648–657. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.; Wang, Z.Y.; Mora-Garcia, S.; Li, J.; Yoshida, S.; Asami, T.; Chory, J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 2002, 109, 181–191. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Nam, K.H. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science 2002, 295, 1299–1301. [Google Scholar]
- Tang, W.; Yuan, M.; Wang, R.; Yang, Y.; Wang, C.; Oses-Prieto, J.A.; Kim, T.W.; Zhou, H.W.; Deng, Z.; Gampala, S.S.; et al. PP2A activates brassinosteroid-responsive gene expression and plant growth by dephosphorylating BZR1. Nat. Cell Biol. 2011, 13, 124–131. [Google Scholar] [CrossRef] [PubMed]
- He, J.X.; Gendron, J.M.; Sun, Y.; Gampala, S.S.; Gendron, N.; Sun, C.Q.; Wang, Z.Y. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 2005, 307, 1634–1638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, Y.; Vafeados, D.; Tao, Y.; Yoshida, S.; Asami, T.; Chory, J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 2005, 120, 249–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peres, A.; Soares, J.S.; Tavares, R.G.; Righetto, G.; Zullo, M.A.T.; Mandava, N.B.; Menossi, M. Brassinosteroids, the sixth class of phytohormones: A molecular view from the discovery to hormonal interactions in plant development and stress adaptation. Int. J. Mol. Sci. 2019, 20, 331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiu, S.H.; Bleecker, A.B. Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci. STKE 2001, 113, re22. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Cano-Delgado, A.; Seto, H.; Hiranuma, S.; Fujioka, S.; Yoshida, S.; Chory, J. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 2005, 433, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Clay, N.K.; Nelson, T. VH1, a provascular cell-specific receptor kinase that influences leaf cell patterns in Arabidopsis. Plant Cell 2002, 14, 2707–2722. [Google Scholar] [CrossRef]
- Cano-Delgado, A.; Yin, Y.; Yu, C.; Vafeados, D.; Mora-Garcia, S.; Cheng, J.C.; Nam, K.H.; Li, J.; Chory, J. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development 2004, 131, 5341–5351. [Google Scholar] [CrossRef] [Green Version]
- Zhou, A.; Wang, H.; Walker, J.C.; Li, J. BRL1, a leucine-rich repeat receptor-like protein kinase, is functionally redundant with BRI1 in regulating Arabidopsis brassinosteroid signaling. Plant J. 2004, 40, 399–409. [Google Scholar] [CrossRef]
- Ceserani, T.; Trofka, A.; Gandotra, N.; Nelson, T. VH1/BRL2 receptor-like kinase interacts with vascular-specific adaptor proteins VIT and VIK to influence leaf venation. Plant J. 2009, 57, 1000–1014. [Google Scholar] [CrossRef] [Green Version]
- Lozano-Elena, F.; Caño-Delgado, A.I. Emerging roles of vascular brassinosteroid receptors of the BRI1-like family. Curr. Opin. Plant Biol. 2019, 51, 105–113. [Google Scholar] [CrossRef]
- Fàbregas, N.; Lozano-Elena, F.; Blasco-Escámez, D.; Tohge, T.; Martínez-Andújar, C.; Albacete, A.; Osorio, S.; Bustamante, M.; Riechmann, J.L.; Nomura, T.; et al. Overexpression of the vascular brassinosteroid receptor BRL3 confers drought resistance without penalizing plant growth. Nat. Commun. 2018, 9, 4680. [Google Scholar] [CrossRef] [Green Version]
- Klok, E.J.; Wilson, I.W.; Wilson, D.; Chapman, S.C.; Ewing, R.M.; Somerville, S.C.; Peacock, W.J.; Dolferus, R.; Dennis, E.S. Expression profile analysis of the low-oxygen response in Arabidopsis root cultures. Plant Cell 2002, 14, 2481–2494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohashi-Ito, K.; Kubo, M.; Demura, T.; Fukuda, H. Class III homeodomain leucine-zipper proteins regulate xylem cell differentiation. Plant Cell Physiol. 2005, 46, 1646–1656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
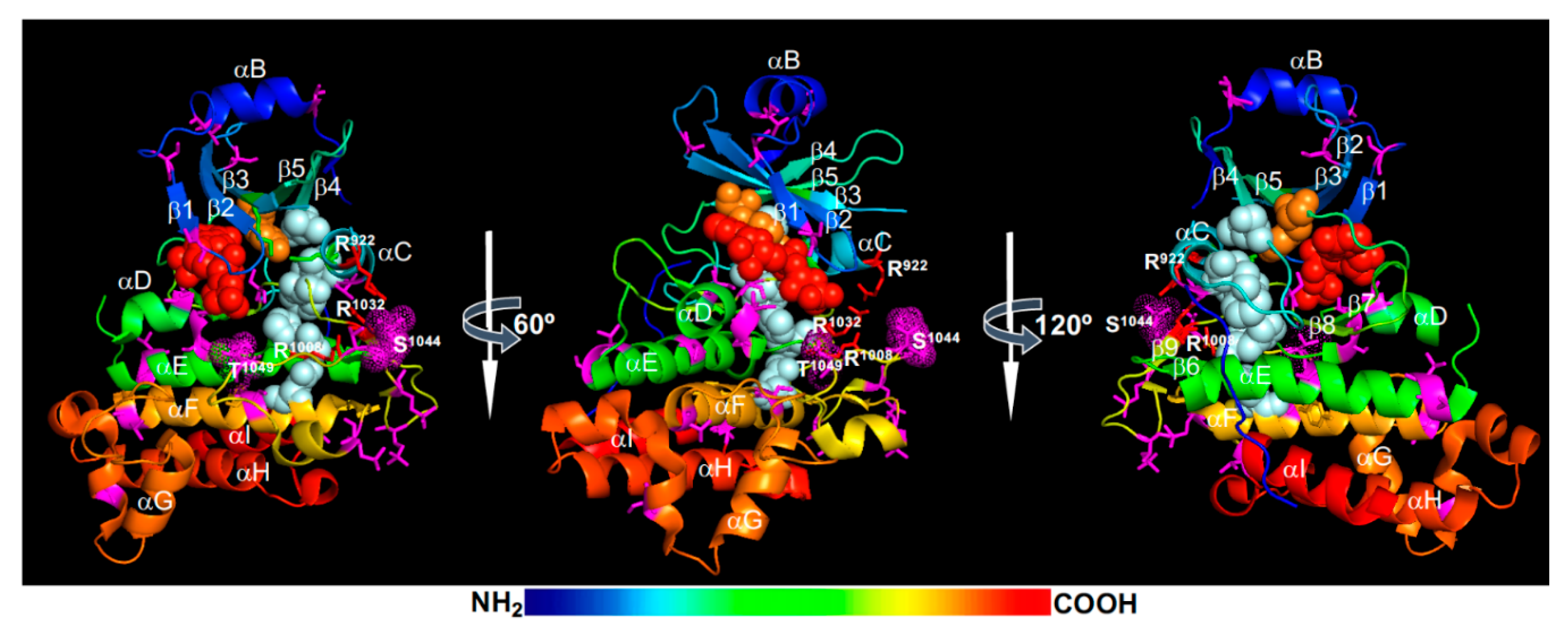
- Wang, J.; Jiang, J.; Wang, J.; Chen, L.; Fan, S.L.; Wu, J.W.; Wang, X.; Wang, Z.X. Structural insights into the negative regulation of BRI1 signaling by BRI1-interacting protein BKI1. Cell Res. 2014, 24, 1328–1341. [Google Scholar] [CrossRef] [Green Version]
- Bojar, D.; Martinez, J.; Santiago, J.; Rybin, V.; Bayliss, R.; Hothorn, M. Crystal structures of the phosphorylated BRI1 kinase domain and implications for brassinosteroid signal initiation. Plant J. 2014, 78, 31–43. [Google Scholar] [CrossRef]
- Bucherl, C.A.; van Esse, G.W.; Kruis, A.; Luchtenberg, J.; Westphal, A.H.; Aker, J.; van Hoek, A.; Albrecht, C.; Borst, J.W.; de Vries, S.C. Visualization of BRI1 and BAK1(SERK3) membrane receptor heterooligomers during brassinosteroid signaling. Plant Physiol. 2013, 162, 1911–1925. [Google Scholar] [CrossRef] [Green Version]
- Hutten, S.J.; Hamers, D.S.; den Toorn, M.A.; van Esse, W.; Nolles, A.; Bucherl, C.A.; de Vries, S.C.; Hohlbein, J.; Borst, J.W. Visualization of BRI1 and SERK3/BAK1 nanoclusters in arabidopsis roots. PLoS ONE 2017, 12, e0169905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gou, X.; Yin, H.; He, K.; Du, J.; Yi, J.; Xu, S.; Lin, H.; Clouse, S.D.; Li, J. Genetic evidence for an indispensable role of somatic embryogenesis receptor kinases in brassinosteroid signaling. PLoS Genet. 2012, 8, e1002452. [Google Scholar] [CrossRef] [Green Version]
- Imkampe, J.; Halter, T.; Huang, S.; Schulze, S.; Mazzotta, S.; Schmidt, N.; Manstretta, R.; Postel, S.; Wierzba, M.; Yang, Y.; et al. The Arabidopsis leucine-rich repeat receptor kinase BIR3 negatively regulates BAK1 receptor complex formation and stabilizes BAK1. Plant Cell 2017, 29, 2285–2303. [Google Scholar] [CrossRef] [Green Version]
- Johnson, L.N.; Noble, M.E.; Owen, D.J. Active and inactive protein kinases: Structural basis for regulation. Cell 1996, 85, 149–158. [Google Scholar] [CrossRef] [Green Version]
- Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 2000, 103, 211–225. [Google Scholar] [CrossRef] [Green Version]
- Heldin, C.H.; Moustakas, A. Signaling receptors for TGF-beta family members. Cold Spring Harb. Perspect. Biol. 2016. [Google Scholar] [CrossRef] [Green Version]
- Hink, M.A.; Shah, K.; Russinova, E.; de Vries, S.C.; Visser, A.J. Fluorescence fluctuation analysis of Arabidopsis thaliana somatic embryogenesis receptor-like kinase and BRASSINOSTEROID INSENSITIVE 1 receptor oligomerization. Biophys. J. 2008, 94, 1052–1062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Li, H.; Lv, X.; Chen, T.; Li, R.; Xue, Y.; Jiang, J.; Jin, B.; Baluska, F.; Samaj, J.; et al. Spatiotemporal dynamics of the BRI1 receptor and its regulation by membrane microdomains in living arabidopsis cells. Mol. Plant 2015, 8, 1334–1349. [Google Scholar] [CrossRef] [Green Version]
- Hohmann, U.; Ramakrishna, P.; Wang, K.; Lorenzo-Orts, L.; Nicolet, J.; Henschen, A.; Barberon, M.; Bayer, M.; Hothorn, M. Constitutive activation of leucine-rich repeat receptor kinase signaling pathways by BAK1-interacting receptor-like kinase 3 chimera. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- She, J.; Han, Z.; Kim, T.W.; Wang, J.; Cheng, W.; Chang, J.; Shi, S.; Wang, J.; Yang, M.; Wang, Z.Y.; et al. Structural insight into brassinosteroid perception by BRI1. Nature 2011, 474, 472–476. [Google Scholar] [CrossRef] [Green Version]
- Hothorn, M.; Belkhadir, Y.; Dreux, M.; Dabi, T.; Noel, J.P.; Wilson, I.A.; Chory, J. Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 2011, 474, 467–471. [Google Scholar] [CrossRef] [Green Version]
- Yan, L.; Ma, Y.; Liu, D.; Wei, X.; Sun, Y.; Chen, X.; Zhao, H.; Zhou, J.; Wang, Z.; Shui, W.; et al. Structural basis for the impact of phosphorylation on the activation of plant receptor-like kinase BAK1. Cell Res. 2012, 22, 1304–1308. [Google Scholar] [CrossRef] [Green Version]
- Grosseholz, R.; Feldman-Salit, A.; Wanke, F.; Schulze, S.; Glockner, N.; Kemmerling, B.; Harter, K.; Kummer, U. Specifying the role of BAK1-interacting receptor-like kinase 3 in brassinosteroid signaling. J. Integr. Plant Biol. 2020, 62, 456–469. [Google Scholar] [CrossRef] [Green Version]
- Kornev, A.P.; Taylor, S.S. Dynamics-driven allostery in protein kinases. Trends Biochem. Sci. 2015, 40, 628–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kornev, A.P.; Haste, N.M.; Taylor, S.S.; Eyck, L.F. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc. Natl. Acad. Sci. USA 2006, 103, 17783–17788. [Google Scholar] [CrossRef] [Green Version]
- Mitra, S.K.; Chen, R.; Dhandaydham, M.; Wang, X.; Blackburn, R.K.; Kota, U.; Goshe, M.B.; Schwartz, D.; Huber, S.C.; Clouse, S.D. An autophosphorylation site database for leucine-rich repeat receptor-like kinases in Arabidopsis thaliana. Plant J. 2015, 82, 1042–1060. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.H.; Ray, W.K.; Huber, S.C.; Asara, J.M.; Gage, D.A.; Clouse, S.D. Recombinant BRASSINOSTEROID INSENSITIVE 1 receptor-like kinase autophosphorylates on serine and threonine residues and phosphorylates a conserved peptide motif in vitro. Plant Physiol. 2000, 124, 751–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, X.; Oh, M.H.; Kim, H.S.; Schwartz, D.; Imai, B.S.; Yau, P.M.; Clouse, S.D.; Huber, S.C. Transphosphorylation of E. coli proteins during production of recombinant protein kinases provides a robust system to characterize kinase specificity. Front. Plant Sci. 2012, 3, 262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Liu, M.; Yuan, M.; Oses-Prieto, J.A.; Cai, X.; Sun, Y.; Burlingame, A.L.; Wang, Z.Y.; Tang, W. The brassinosteroid-activated BRI1 receptor kinase is switched off by dephosphorylation mediated by cytoplasm-localized PP2A B’ subunits. Mol. Plant 2016, 9, 148–157. [Google Scholar] [CrossRef]
- Oh, M.H.; Wang, X.; Kota, U.; Goshe, M.B.; Clouse, S.D.; Huber, S.C. Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 658–663. [Google Scholar] [CrossRef] [Green Version]
- Oh, M.H.; Clouse, S.D.; Huber, S.C. Tyrosine phosphorylation of the BRI1 receptor kinase occurs via a post-translational modification and is activated by the juxtamembrane domain. Front. Plant Sci. 2012, 3, 175. [Google Scholar] [CrossRef] [Green Version]
- Amorim-Silva, V.; Garcia-Moreno, A.; Castillo, A.G.; Lakhssassi, N.; Del Valle, A.E.; Perez-Sancho, J.; Li, Y.; Pose, D.; Perez-Rodriguez, J.; Lin, J.; et al. TTL proteins scaffold brassinosteroid signaling components at the plasma membrane to optimize signal transduction in arabidopsis. Plant Cell 2019, 31, 1807–1828. [Google Scholar] [CrossRef] [Green Version]
- Ren, H.; Willige, B.C.; Jaillais, Y.; Geng, S.; Park, M.Y.; Gray, W.M.; Chory, J. Brassinosteroid-signaling kinase 3, a plasma membrane-associated scaffold protein involved in early brassinosteroid signaling. PLoS Genet. 2019, 15, e1007904. [Google Scholar] [CrossRef] [Green Version]
- Jaillais, Y.; Hothorn, M.; Belkhadir, Y.; Dabi, T.; Nimchuk, Z.L.; Meyerowitz, E.M.; Chory, J. Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 2011, 25, 232–237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Yang, C.; Zhang, C.; Wang, N.; Lu, D.; Wang, J.; Zhang, S.; Wang, Z.X.; Ma, H.; Wang, X. Dual role of BKI1 and 14-3-3 s in brassinosteroid signaling to link receptor with transcription factors. Dev. Cell 2011, 21, 825–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, M.H.; Wang, X.; Clouse, S.D.; Huber, S.C. Deactivation of the Arabidopsis BRASSINOSTEROID INSENSITIVE 1 (BRI1) receptor kinase by autophosphorylation within the glycine-rich loop. Proc. Natl. Acad. Sci. USA 2012, 109, 327–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.; Wang, X.; Li, X.; Kamiya, Y.; Otegui, M.S.; Chory, J. Methylation of a phosphatase specifies dephosphorylation and degradation of activated brassinosteroid receptors. Sci. Signal. 2011, 4, ra29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janssens, V.; Goris, J. Protein phosphatase 2A: A highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 2001, 353, 417–439. [Google Scholar] [CrossRef]
- Macho, A.P.; Schwessinger, B.; Ntoukakis, V.; Brutus, A.; Segonzac, C.; Roy, S.; Kadota, Y.; Oh, M.H.; Sklenar, J.; Derbyshire, P.; et al. A bacterial tyrosine phosphatase inhibits plant pattern recognition receptor activation. Science 2014, 343, 1509–1512. [Google Scholar] [CrossRef]
- Kerk, D.; Templeton, G.; Moorhead, G.B. Evolutionary radiation pattern of novel protein phosphatases revealed by analysis of protein data from the completely sequenced genomes of humans, green algae, and higher plants. Plant Physiol. 2008, 146, 351–367. [Google Scholar] [CrossRef] [Green Version]
- Araki, K.; Nagata, K. Protein folding and quality control in the ER. Cold Spring Harb. Perspect. Biol. 2011, 3, a007526. [Google Scholar] [CrossRef] [Green Version]
- Strasser, R. Protein quality control in the endoplasmic reticulum of plants. Annu. Rev. Plant Biol. 2018, 69, 147–172. [Google Scholar] [CrossRef]
- Jin, H.; Hong, Z.; Su, W.; Li, J. A plant-specific calreticulin is a key retention factor for a defective brassinosteroid receptor in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2009, 106, 13612–13617. [Google Scholar] [CrossRef] [Green Version]
- Jin, H.; Yan, Z.; Nam, K.H.; Li, J. Allele-specific suppression of a defective brassinosteroid receptor reveals a physiological role of UGGT in ER quality control. Mol. Cell 2007, 26, 821–830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, Z.; Jin, H.; Tzfira, T.; Li, J. Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 2008, 20, 3418–3429. [Google Scholar] [CrossRef] [Green Version]
- Su, W.; Liu, Y.; Xia, Y.; Hong, Z.; Li, J. Conserved endoplasmic reticulum-associated degradation system to eliminate mutated receptor-like kinases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 870–875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, W.; Liu, Y.; Xia, Y.; Hong, Z.; Li, J. The Arabidopsis homolog of the mammalian OS-9 protein plays a key role in the endoplasmic reticulum-associated degradation of misfolded receptor-like kinases. Mol. Plant 2012, 5, 929–940. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.; Zhang, C.; Chen, Y.; Wang, Y.; Wang, D.; Liu, X.; Wang, M.; Mao, J.; Zhang, J.; Xing, W.; et al. PAWH1 and PAWH2 are plant-specific components of an Arabidopsis endoplasmic reticulum-associated degradation complex. Nat. Commun. 2019, 10, 3492. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, C.; Wang, D.; Su, W.; Liu, L.; Wang, M.; Li, J. EBS7 is a plant-specific component of a highly conserved endoplasmic reticulum-associated degradation system in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, 12205–12210. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zhou, L.; Qin, Y.; Chen, Y.; Liu, X.; Wang, M.; Mao, J.; Zhang, J.; He, Z.; Liu, L.; et al. A temperature-sensitive misfolded bri1-301 receptor requires its kinase activity to promote growth. Plant Physiol. 2018, 178, 1704–1719. [Google Scholar] [CrossRef] [Green Version]
- Lv, M.; Li, M.; Chen, W.; Wang, Y.; Sun, C.; Yin, H.; He, K.; Li, J. Thermal-enhanced bri1-301 instability reveals a plasma membrane protein quality control system in plants. Front. Plant Sci. 2018, 9, 1620. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Otegui, M.S.; Spalding, E.P. The ER-localized TWD1 immunophilin is necessary for localization of multidrug resistance-like proteins required for polar auxin transport in Arabidopsis roots. Plant Cell 2010, 22, 3295–3304. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.; Lv, M.; Feng, Z.; Campbell, T.; Liscum, E.; Li, J. TWISTED DWARF 1 associates with BRASSINOSTEROID-INSENSITIVE 1 to regulate early events of the brassinosteroid signaling pathway. Mol. Plant 2016, 9, 582–592. [Google Scholar] [CrossRef] [Green Version]
- Chaiwanon, J.; Garcia, V.J.; Cartwright, H.; Sun, Y.; Wang, Z.Y. Immunophilin-like FKBP42/TWISTED DWARF1 interacts with the receptor kinase BRI1 to regulate brassinosteroid signaling in Arabidopsis. Mol. Plant 2016, 9, 593–600. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geisler, M.; Kolukisaoglu, H.U.; Bouchard, R.; Billion, K.; Berger, J.; Saal, B.; Frangne, N.; Koncz-Kalman, Z.; Koncz, C.; Dudler, R.; et al. TWISTED DWARF1, a unique plasma membrane-anchored immunophilin-like protein, interacts with Arabidopsis multidrug resistance-like transporters AtPGP1 and AtPGP19. Mol. Biol. Cell 2003, 14, 4238–4249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.; Bailly, A.; Zwiewka, M.; Sovero, V.; Di Donato, M.; Ge, P.; Oehri, J.; Aryal, B.; Hao, P.; Linnert, M.; et al. TWISTED DWARF1 mediates the action of auxin transport inhibitors on actin cytoskeleton dynamics. Plant Cell 2016, 28, 930–948. [Google Scholar] [CrossRef] [Green Version]
- Ott, T. Membrane nanodomains and microdomains in plant-microbe interactions. Curr. Opin. Plant Biol. 2017, 40, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Bucherl, C.A.; Jarsch, I.K.; Schudoma, C.; Segonzac, C.; Mbengue, M.; Robatzek, S.; MacLean, D.; Ott, T.; Zipfel, C. Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. Elife 2017, 6, e25114. [Google Scholar] [CrossRef] [PubMed]
- Gui, J.; Zheng, S.; Liu, C.; Shen, J.; Li, J.; Li, L. OsREM4.1 interacts with OsSERK1 to coordinate the interlinking between abscisic acid and brassinosteroid signaling in rice. Dev. Cell 2016, 38, 201–213. [Google Scholar] [CrossRef] [Green Version]
- Jaillais, Y.; Ott, T. The nanoscale organization of the plasma membrane and its importance in signaling: A proteolipid perspective. Plant Physiol. 2020, 182, 1682–1696. [Google Scholar] [CrossRef] [Green Version]
- Shiu, S.H.; Karlowski, W.M.; Pan, R.; Tzeng, Y.H.; Mayer, K.F.; Li, W.H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar] [CrossRef] [Green Version]
- Geldner, N.; Hyman, D.L.; Wang, X.; Schumacher, K.; Chory, J. Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 2007, 21, 1598–1602. [Google Scholar] [CrossRef] [Green Version]
- Claus, L.A.N.; Savatin, D.V.; Russinova, E. The crossroads of receptor-mediated signaling and endocytosis in plants. J. Integr. Plant Biol. 2018, 60, 827–840. [Google Scholar] [CrossRef] [Green Version]
- Dettmer, J.; Hong-Hermesdorf, A.; Stierhof, Y.D.; Schumacher, K. Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 2006, 18, 715–730. [Google Scholar] [CrossRef] [Green Version]
- Viotti, C.; Bubeck, J.; Stierhof, Y.D.; Krebs, M.; Langhans, M.; van den Berg, W.; van Dongen, W.; Richter, S.; Geldner, N.; Takano, J.; et al. Endocytic and secretory traffic in Arabidopsis merge in the trans-Golgi network/early endosome, an independent and highly dynamic organelle. Plant Cell 2010, 22, 1344–1357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaillais, Y.; Fobis-Loisy, I.; Miege, C.; Gaude, T. Evidence for a sorting endosome in Arabidopsis root cells. Plant J. 2008, 53, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Irani, N.G.; Di Rubbo, S.; Mylle, E.; van den Begin, J.; Schneider-Pizon, J.; Hnilikova, J.; Sisa, M.; Buyst, D.; Vilarrasa-Blasi, J.; Szatmari, A.M.; et al. Fluorescent castasterone reveals BRI1 signaling from the plasma membrane. Nat. Chem. Biol. 2012, 8, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Geldner, N.; Robatzek, S. Plant receptors go endosomal: A moving view on signal transduction. Plant Physiol. 2008, 147, 1565–1574. [Google Scholar] [CrossRef] [Green Version]
- Reynolds, G.D.; Wang, C.; Pan, J.; Bednarek, S.Y. Inroads into internalization: Five years of endocytic exploration. Plant Physiol. 2018, 176, 208–218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banbury, D.N.; Oakley, J.D.; Sessions, R.B.; Banting, G. Tyrphostin A23 inhibits internalization of the transferrin receptor by perturbing the interaction between tyrosine motifs and the medium chain subunit of the AP-2 adaptor complex. J. Biol. Chem. 2003, 278, 12022–12028. [Google Scholar] [CrossRef] [Green Version]
- Gadeyne, A.; Sánchez-Rodríguez, C.; Vanneste, S.; Di Rubbo, S.; Zauber, H.; Vanneste, K.; van Leene, J.; de Winne, N.; Eeckhout, D.; Persiau, G.; et al. The TPLATE adaptor complex drives clathrin-mediated endocytosis in plants. Cell 2014, 156, 691–704. [Google Scholar] [CrossRef] [Green Version]
- Di Rubbo, S.; Irani, N.G.; Kim, S.Y.; Xu, Z.Y.; Gadeyne, A.; Dejonghe, W.; Vanhoutte, I.; Persiau, G.; Eeckhout, D.; Simon, S.; et al. The clathrin adaptor complex AP-2 mediates endocytosis of brassinosteroid insensitive1 in Arabidopsis. Plant Cell 2013, 25, 2986–2997. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Liu, P.; Wan, Y.; Chen, T.; Wang, Q.; Mettbach, U.; Baluska, F.; Samaj, J.; Fang, X.; Lucas, W.J.; et al. A membrane microdomain-associated protein, Arabidopsis Flot1, is involved in a clathrin-independent endocytic pathway and is required for seedling development. Plant Cell 2012, 24, 2105–2122. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Furlan, C.; Minina, E.A.; Hicks, G.R. Remove, recycle, degrade: Regulating plasma membrane protein accumulation. Plant Cell 2019, 31, 2833–2854. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liu, D.; Wang, P.; Ma, X.; Lin, W.; Chen, S.; Mishev, K.; Lu, D.; Kumar, R.; Vanhoutte, I.; et al. Regulation of Arabidopsis brassinosteroid receptor BRI1 endocytosis and degradation by plant U-box PUB12/PUB13-mediated ubiquitination. Proc. Natl. Acad. Sci. USA 2018, 115, E1906–E1915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardona-Lopez, X.; Cuyas, L.; Marin, E.; Rajulu, C.; Irigoyen, M.L.; Gil, E.; Puga, M.I.; Bligny, R.; Nussaume, L.; Geldner, N.; et al. ESCRT-III-associated protein ALIX mediates high-affinity phosphate transporter trafficking to maintain phosphate homeostasis in arabidopsis. Plant Cell 2015, 27, 2560–2581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mattern, M.; Sutherland, J.; Kadimisetty, K.; Barrio, R.; Rodriguez, M.S. Using ubiquitin binders to decipher the ubiquitin code. Trends. Biochem. Sci. 2019, 44, 599–615. [Google Scholar] [CrossRef] [PubMed]
- Martins, S.; Dohmann, E.M.; Cayrel, A.; Johnson, A.; Fischer, W.; Pojer, F.; Satiat-Jeunemaitre, B.; Jaillais, Y.; Chory, J.; Geldner, N.; et al. Internalization and vacuolar targeting of the brassinosteroid hormone receptor BRI1 are regulated by ubiquitination. Nat. Commun. 2015, 6, 6151. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Gomez, L.; Boller, T. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol. Cell 2000, 5, 1003–1011. [Google Scholar] [CrossRef]
- Lu, D.; Lin, W.; Gao, X.; Wu, S.; Cheng, C.; Avila, J.; Heese, A.; Devarenne, T.P.; He, P.; Shan, L. Direct ubiquitination of pattern recognition receptor FLS2 attenuates plant innate immunity. Science 2011, 332, 1439–1442. [Google Scholar] [CrossRef] [Green Version]
- Kalinowska, K.; Nagel, M.K.; Goodman, K.; Cuyas, L.; Anzenberger, F.; Alkofer, A.; Paz-Ares, J.; Braun, P.; Rubio, V.; Otegui, M.S.; et al. Arabidopsis ALIX is required for the endosomal localization of the deubiquitinating enzyme AMSH3. Proc. Natl. Acad. Sci. USA 2015, 112, E5543–E5551. [Google Scholar] [CrossRef] [Green Version]
- Yamagami, A.; Saito, C.; Nakazawa, M.; Fujioka, S.; Uemura, T.; Matsui, M.; Sakuta, M.; Shinozaki, K.; Osada, H.; Nakano, A.; et al. Evolutionarily conserved BIL4 suppresses the degradation of brassinosteroid receptor BRI1 and regulates cell elongation. Sci. Rep. 2017, 7, 5739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Y.; Scholl, S.; Doering, A.; Zhang, Y.; Irani, N.G.; Rubbo, S.D.; Neumetzler, L.; Krishnamoorthy, P.; Van Houtte, I.; Mylle, E.; et al. V-ATPase activity in the TGN/EE is required for exocytosis and recycling in Arabidopsis. Nat. Plants 2015, 1, 15094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schumacher, K.; Vafeados, D.; McCarthy, M.; Sze, H.; Wilkins, T.; Chory, J. The Arabidopsis det3 mutant reveals a central role for the vacuolar H(+)-ATPase in plant growth and development. Genes Dev. 1999, 13, 3259–3270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ambrose, C.; Ruan, Y.; Gardiner, J.; Tamblyn, L.M.; Catching, A.; Kirik, V.; Marc, J.; Overall, R.; Wasteneys, G.O. CLASP interacts with sorting nexin 1 to link microtubules and auxin transport via PIN2 recycling in Arabidopsis thaliana. Dev. Cell 2013, 24, 649–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruan, Y.; Halat, L.S.; Khan, D.; Jancowski, S.; Ambrose, C.; Belmonte, M.F.; Wasteneys, G.O. The microtubule-associated protein CLASP sustains cell proliferation through a brassinosteroid signaling negative feedback loop. Curr. Biol. 2018, 28, 2718–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Xing, J.; Lin, J. At the intersection of exocytosis and endocytosis in plants. New Phytol. 2019, 224, 1479–1489. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, Y.; Zhu, X.F.; Jung, J.H.; Sun, Q.; Li, T.Y.; Chen, L.J.; Duan, Y.X.; Xuan, Y.H. SYP22 and VAMP727 regulate BRI1 plasma membrane targeting to control plant growth in Arabidopsis. New Phytol. 2019, 223, 1059–1065. [Google Scholar] [CrossRef] [Green Version]
- Drdova, E.J.; Synek, L.; Pecenkova, T.; Hala, M.; Kulich, I.; Fowler, J.E.; Murphy, A.S.; Zarsky, V. The exocyst complex contributes to PIN auxin efflux carrier recycling and polar auxin transport in Arabidopsis. Plant J. 2013, 73, 709–719. [Google Scholar] [CrossRef]
- Xue, S.; Zou, J.; Liu, Y.; Wang, M.; Zhang, C.; Le, J. Involvement of BIG5 and BIG3 in BRI1 trafficking reveals diverse functions of BIG-subfamily ARF-GEFs in plant growth and gravitropism. Int. J. Mol. Sci. 2019, 20, 2339. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Wu, Y.; Gao, Y.; Li, M.; Yin, H.; Lv, M.; Zhao, J.; Li, J.; He, K. Somatic embryogenesis receptor-like kinase 5 in the ecotype Landsberg erecta of Arabidopsis is a functional RD LRR-RLK in regulating brassinosteroid signaling and cell death control. Front Plant Sci. 2015, 6, 852. [Google Scholar] [CrossRef] [Green Version]
- Chinchilla, D.; Zipfel, C.; Robatzek, S.; Kemmerling, B.; Nürnberger, T.; Jones, J.D.G.; Felix, G.; Boller, T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature 2007, 448, 497–500. [Google Scholar] [CrossRef]
- Karlova, R.; Boeren, S.; Russinova, E.; Aker, J.; Vervoort, J.; de Vries, S. The arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE1 protein complex includes BRASSINOSTEROID-INSENSITIVE1. Plant Cell 2006, 18, 626–638. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Liu, Y.; Xu, F.; Zhang, Y. Differential requirement of BAK1 C-terminal tail in development and immunity. J. Integr. Plant Biol. 2018, 60, 270–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Li, Z.; Liu, D.; Xu, J.; Wei, X.; Yan, L.; Yang, C.; Lou, Z.; Shui, W. Assessment of BAK1 activity in different plant receptor-like kinase complexes by quantitative profiling of phosphorylation patterns. J. Proteomics 2014, 108, 484–493. [Google Scholar] [CrossRef] [PubMed]
- Shah, K.; Vervoort, J.; de Vries, S.C. Role of threonines in the Arabidopsis thaliana somatic embryogenesis receptor kinase 1 activation loop in phosphorylation. J. Biol. Chem. 2001, 276, 41263–41269. [Google Scholar] [CrossRef] [Green Version]
- Perraki, A.; DeFalco, T.A.; Derbyshire, P.; Avila, J.; Sere, D.; Sklenar, J.; Qi, X.; Stransfeld, L.; Schwessinger, B.; Kadota, Y.; et al. Phosphocode-dependent functional dichotomy of a common co-receptor in plant signalling. Nature 2018, 561, 248–252. [Google Scholar] [CrossRef]
- Singh, V.; Perraki, A.; Kim, S.Y.; Shrivastava, S.; Lee, J.H.; Zhao, Y.; Schwessinger, B.; Oh, M.H.; Marshall-Colon, A.; Zipfel, C.; et al. Tyrosine-610 in the receptor kinase BAK1 does not play a major role in brassinosteroid signaling or innate immunity. Front Plant Sci. 2017, 8, 1273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, W.; Li, B.; Lu, D.; Chen, S.; Zhu, N.; He, P.; Shan, L. Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity. Proc. Natl. Acad. Sci. USA 2014, 111, 3632–3637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segonzac, C.; Macho, A.P.; Sanmartin, M.; Ntoukakis, V.; Sanchez-Serrano, J.J.; Zipfel, C. Negative control of BAK1 by protein phosphatase 2A during plant innate immunity. EMBO J. 2014, 33, 2069–2079. [Google Scholar] [CrossRef] [Green Version]
- Song, L.; Shi, Q.M.; Yang, X.H.; Xu, Z.H.; Xue, H.W. Membrane steroid-binding protein 1 (MSBP1) negatively regulates brassinosteroid signaling by enhancing the endocytosis of BAK1. Cell Res. 2009, 19, 864–876. [Google Scholar] [CrossRef]
- Gao, M.; Wang, X.; Wang, D.; Xu, F.; Ding, X.; Zhang, Z.; Bi, D.; Cheng, Y.T.; Chen, S.; Li, X.; et al. Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 2009, 6, 34–44. [Google Scholar] [CrossRef] [Green Version]
- Halter, T.; Imkampe, J.; Mazzotta, S.; Wierzba, M.; Postel, S.; Bucherl, C.; Kiefer, C.; Stahl, M.; Chinchilla, D.; Wang, X.; et al. The leucine-rich repeat receptor kinase BIR2 is a negative regulator of BAK1 in plant immunity. Curr. Biol. 2014, 24, 134–143. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Huang, X.; Li, M.; He, P.; Zhang, Y. Loss-of-function of Arabidopsis receptor-like kinase BIR1 activates cell death and defense responses mediated by BAK1 and SOBIR1. New Phytol. 2016, 212, 637–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halter, T.; Imkampe, J.; Blaum, B.S.; Stehle, T.; Kemmerling, B. BIR2 affects complex formation of BAK1 with ligand binding receptors in plant defense. Plant Signal. Behav. 2014, 9, e28944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hohmann, U.; Nicolet, J.; Moretti, A.; Hothorn, L.A.; Hothorn, M. The SERK3 elongated allele defines a role for BIR ectodomains in brassinosteroid signalling. Nat. Plants 2018, 4, 345–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, Y.; Choe, V.; Fujioka, S.; Takatsuto, S.; Han, M.; Jeon, J.S.; Park, Y.I.; Lee, K.O.; Choe, S. Constitutive activation of brassinosteroid signaling in the Arabidopsis elongated-D/bak1 mutant. Plant Mol. Biol. 2012, 80, 489–501. [Google Scholar] [CrossRef]
- Halliday, K.; Devlin, P.F.; Whitelam, G.C.; Hanhart, C.; Koornneef, M. The ELONGATED gene of Arabidopsis acts independently of light and gibberellins in the control of elongation growth. Plant J. 1996, 9, 305–312. [Google Scholar] [CrossRef]
- Kim, M.H.; Kim, Y.; Kim, J.W.; Lee, H.S.; Lee, W.S.; Kim, S.K.; Wang, Z.Y.; Kim, S.H. Identification of Arabidopsis BAK1-associating receptor-like kinase 1 (BARK1) and characterization of its gene expression and brassinosteroid-regulated root phenotypes. Plant Cell Physiol. 2013, 54, 1620–1634. [Google Scholar] [CrossRef] [Green Version]
- Blaum, B.S.; Mazzotta, S.; Noldeke, E.R.; Halter, T.; Madlung, J.; Kemmerling, B.; Stehle, T. Structure of the pseudokinase domain of BIR2, a regulator of BAK1-mediated immune signaling in Arabidopsis. J. Struct. Biol. 2014, 186, 112–121. [Google Scholar] [CrossRef]
- Grek, C.L.; Zhang, J.; Manevich, Y.; Townsend, D.M.; Tew, K.D. Causes and consequences of cysteine S-glutathionylation. J. Biol. Chem. 2013, 288, 26497–26504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bender, K.W.; Wang, X.; Cheng, G.B.; Kim, H.S.; Zielinski, R.E.; Huber, S.C. Glutaredoxin AtGRXC2 catalyses inhibitory glutathionylation of Arabidopsis BRI1-associated receptor-like kinase 1 (BAK1) in vitro. Biochem. J. 2015, 467, 399–413. [Google Scholar] [CrossRef] [PubMed]
- Moffett, A.S.; Bender, K.W.; Huber, S.C.; Shukla, D. Allosteric control of a plant receptor kinase through s-glutathionylation. Biophys. J. 2017, 113, 2354–2363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwessinger, B.; Roux, M.; Kadota, Y.; Ntoukakis, V.; Sklenar, J.; Jones, A.; Zipfel, C. Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 2011, 7, e1002046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, J.; Wang, P.; Claus, L.A.N.; Savatin, D.V.; Xu, G.; Wu, S.; Meng, X.; Russinova, E.; He, P.; Shan, L. Proteolytic processing of SERK3/BAK1 regulates plant immunity, development, and cell death. Plant Physiol. 2019, 180, 543–558. [Google Scholar] [CrossRef] [Green Version]
- Ntoukakis, V.; Schwessinger, B.; Segonzac, C.; Zipfel, C. Cautionary notes on the use of C-terminal BAK1 fusion proteins for functional studies. Plant Cell 2011, 23, 3871–3878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, D.; Wu, S.; Gao, X.; Zhang, Y.; Shan, L.; He, P. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 2010, 107, 496–501. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Peng, P.; Schmitz, R.J.; Decker, A.D.; Tax, F.E.; Li, J. Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol. 2002, 130, 1221–1229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.Y.; Nakano, T.; Gendron, J.; He, J.; Chen, M.; Vafeados, D.; Yang, Y.; Fujioka, S.; Yoshida, S.; Asami, T.; et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell 2002, 2, 505–513. [Google Scholar] [CrossRef] [Green Version]
- Youn, J.H.; Kim, T.W. Functional insights of plant GSK3-like kinases: Multi-taskers in diverse cellular signal transduction pathways. Mol. Plant 2015, 8, 552–565. [Google Scholar] [CrossRef] [Green Version]
- Choe, S.; Schmitz, R.J.; Fujioka, S.; Takatsuto, S.; Lee, M.O.; Yoshida, S.; Feldmann, K.A.; Tax, F.E. Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semidominant and defective in a glycogen synthase kinase 3beta-like kinase. Plant Physiol. 2002, 130, 1506–1515. [Google Scholar] [CrossRef] [Green Version]
- Perez-Perez, J.M.; Ponce, M.R.; Micol, J.L. The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev. Biol. 2002, 242, 161–173. [Google Scholar] [CrossRef] [Green Version]
- Yan, Z.; Zhao, J.; Peng, P.; Chihara, R.K.; Li, J. BIN2 functions redundantly with other Arabidopsis GSK3-like kinases to regulate brassinosteroid signaling. Plant Physiol. 2009, 150, 710–721. [Google Scholar] [CrossRef] [Green Version]
- Youn, J.H.; Kim, T.W.; Kim, E.J.; Bu, S.; Kim, S.K.; Wang, Z.Y.; Kim, T.W. Structural and functional characterization of Arabidopsis GSK3-like kinase AtSK12. Mol. Cells 2013, 36, 564–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Rybel, B.; Audenaert, D.; Vert, G.; Rozhon, W.; Mayerhofer, J.; Peelman, F.; Coutuer, S.; Denayer, T.; Jansen, L.; Nguyen, L.; et al. Chemical inhibition of a subset of Arabidopsis thaliana GSK3-like kinases activates brassinosteroid signaling. Chem. Biol. 2009, 16, 594–604. [Google Scholar] [CrossRef] [PubMed]
- Vert, G.; Chory, J. Downstream nuclear events in brassinosteroid signalling. Nature 2006, 441, 96–100. [Google Scholar] [CrossRef] [PubMed]
- Beurel, E.; Grieco, S.F.; Jope, R.S. Glycogen synthase kinase-3 (GSK3): Regulation, actions, and diseases. Pharmacol. Ther. 2015, 148, 114–131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, P.; Zhao, J.; Zhu, Y.; Asami, T.; Li, J. A direct docking mechanism for a plant GSK3-like kinase to phosphorylate its substrates. J. Biol. Chem. 2010, 285, 24646–24653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Tang, J.; Liu, J.; Hu, J.; Liu, J.; Chen, Y.; Cai, Z.; Wang, X. Abscisic acid signaling inhibits brassinosteroid signaling through dampening the dephosphorylation of BIN2 by ABI1 and ABI2. Mol. Plant 2018, 11, 315–325. [Google Scholar] [CrossRef] [Green Version]
- Lochhead, P.A.; Kinstrie, R.; Sibbet, G.; Rawjee, T.; Morrice, N.; Cleghon, V. A chaperone-dependent GSK3beta transitional intermediate mediates activation-loop autophosphorylation. Mol. Cell 2006, 24, 627–633. [Google Scholar] [CrossRef]
- Cole, A.; Frame, S.; Cohen, P. Further evidence that the tyrosine phosphorylation of glycogen synthase kinase-3 (GSK3) in mammalian cells is an autophosphorylation event. Biochem. J. 2004, 377, 249–255. [Google Scholar] [CrossRef] [Green Version]
- Sugiyama, N.; Nakagami, H.; Mochida, K.; Daudi, A.; Tomita, M.; Shirasu, K.; Ishihama, Y. Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol. Syst. Biol. 2008, 4, 193. [Google Scholar] [CrossRef]
- Lin, L.L.; Hsu, C.L.; Hu, C.W.; Ko, S.Y.; Hsieh, H.L.; Huang, H.C.; Juan, H.F. Integrating phosphoproteomics and bioinformatics to study brassinosteroid-regulated phosphorylation dynamics in arabidopsis. BMC Genomics 2015, 16, 533. [Google Scholar] [CrossRef] [Green Version]
- Hoffmeister, L.; Diekmann, M.; Brand, K.; Huber, R. GSK3: A kinase balancing promotion and resolution of inflammation. Cells 2020, 9, 820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hughes, K.; Nikolakaki, E.; Plyte, S.E.; Totty, N.F.; Woodgett, J.R. Modulation of the glycogen synthase kinase-3 family by tyrosine phosphorylation. EMBO J. 1993, 12, 803–808. [Google Scholar] [CrossRef]
- Dajani, R.; Fraser, E.; Roe, S.M.; Young, N.; Good, V.; Dale, T.C.; Pearl, L.H. Crystal structure of glycogen synthase kinase 3 beta: Structural basis for phosphate-primed substrate specificity and autoinhibition. Cell 2001, 105, 721–732. [Google Scholar] [CrossRef]
- Buescher, J.L.; Phiel, C.J. A noncatalytic domain of glycogen synthase kinase-3 (GSK-3) is essential for activity. J. Biol. Chem. 2010, 285, 7957–7963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itoh, K.; Tang, T.L.; Neel, B.G.; Sokol, S.Y. Specific modulation of ectodermal cell fates in Xenopus embryos by glycogen synthase kinase. Development 1995, 121, 3979–3988. [Google Scholar]
- Moorhead, G.B.; de Wever, V.; Templeton, G.; Kerk, D. Evolution of protein phosphatases in plants and animals. Biochem. J. 2009, 417, 401–409. [Google Scholar] [CrossRef] [Green Version]
- Mora-Garcia, S.; Vert, G.; Yin, Y.; Cano-Delgado, A.; Cheong, H.; Chory, J. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004, 18, 448–460. [Google Scholar] [CrossRef] [Green Version]
- Maselli, G.A.; Slamovits, C.H.; Bianchi, J.I.; Vilarrasa-Blasi, J.; Cano-Delgado, A.I.; Mora-Garcia, S. Revisiting the evolutionary history and roles of protein phosphatases with Kelch-like domains in plants. Plant Physiol. 2014, 164, 1527–1541. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y. Serine/threonine phosphatases: Mechanism through structure. Cell 2009, 139, 468–484. [Google Scholar] [CrossRef] [Green Version]
- Tonks, N.K. Protein tyrosine phosphatases: From genes, to function, to disease. Nat. Rev. Mol. Cell Biol. 2006, 7, 833–846. [Google Scholar] [CrossRef]
- Gao, X.; Zhang, J.Q.; Zhang, X.; Zhou, J.; Jiang, Z.; Huang, P.; Tang, Z.; Bao, Y.; Cheng, J.; Tang, H.; et al. Rice qGL3/OsPPKL1 functions with the GSK3/SHAGGY-like kinase OsGSK3 to modulate brassinosteroid signaling. Plant Cell 2019, 31, 1077–1093. [Google Scholar] [CrossRef] [PubMed]
- Merlot, S.; Gosti, F.; Guerrier, D.; Vavasseur, A.; Giraudat, J. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J. 2001, 25, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Cai, Z.; Wang, X. The primary signaling outputs of brassinosteroids are regulated by abscisic acid signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 4543–4548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, Y.; Wang, H.; Qiao, S.; Leng, L.; Wang, X. Histone deacetylase HDA6 enhances brassinosteroid signaling by inhibiting the BIN2 kinase. Proc. Natl. Acad. Sci. USA 2016, 113, 10418–10423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- ter Haar, E.; Coll, J.T.; Austen, D.A.; Hsiao, H.M.; Swenson, L.; Jain, J. Structure of GSK3beta reveals a primed phosphorylation mechanism. Nat. Struct. Biol. 2001, 8, 593–596. [Google Scholar] [CrossRef]
- Monteserin-Garcia, J.; Al-Massadi, O.; Seoane, L.M.; Alvarez, C.V.; Shan, B.; Stalla, J.; Paez-Pereda, M.; Casanueva, F.F.; Stalla, G.K.; Theodoropoulou, M. Sirt1 inhibits the transcription factor CREB to regulate pituitary growth hormone synthesis. FASEB J. 2013, 27, 1561–1571. [Google Scholar] [CrossRef]
- Taylor, I.; Wang, Y.; Seitz, K.; Baer, J.; Bennewitz, S.; Mooney, B.P.; Walker, J.C. Analysis of phosphorylation of the receptor-like protein kinase HAESA during arabidopsis floral abscission. PLoS ONE 2016, 11, e0147203. [Google Scholar] [CrossRef] [Green Version]
- Sarikhani, M.; Mishra, S.; Maity, S.; Kotyada, C.; Wolfgeher, D.; Gupta, M.P.; Singh, M.; Sundaresan, N.R. SIRT2 deacetylase regulates the activity of GSK3 isoforms independent of inhibitory phosphorylation. Elife 2018, 7, e32925. [Google Scholar] [CrossRef]
- Wang, P.; Du, Y.; Hou, Y.J.; Zhao, Y.; Hsu, C.C.; Yuan, F.; Zhu, X.; Tao, W.A.; Song, C.P.; Zhu, J.K. Nitric oxide negatively regulates abscisic acid signaling in guard cells by S-nitrosylation of OST1. Proc. Natl. Acad. Sci. USA 2015, 112, 613–618. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.B.; Venkatraman, V.; Crowgey, E.L.; Liu, T.; Fu, Z.; Holewinski, R.; Ranek, M.; Kass, D.A.; O’Rourke, B.; van Eyk, J.E. Protein s-nitrosylation controls glycogen synthase kinase 3beta function independent of its phosphorylation state. Circ. Res. 2018, 122, 1517–1531. [Google Scholar] [CrossRef]
- Song, S.; Wang, H.; Sun, M.; Tang, J.; Zheng, B.; Wang, X.; Tan, Y.W. Reactive oxygen species-mediated BIN2 activity revealed by single-molecule analysis. New Phytol. 2019, 223, 692–704. [Google Scholar] [CrossRef] [PubMed]
- He, G.; Liu, J.; Dong, H.; Sun, J. The blue-light receptor CRY1 interacts with BZR1 and BIN2 to modulate the phosphorylation and nuclear function of BZR1 in repressing BR signaling in Arabidopsis. Mol. Plant 2019, 12, 689–703. [Google Scholar] [CrossRef] [PubMed]
- Ling, J.J.; Li, J.; Zhu, D.; Deng, X.W. Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. Proc. Natl. Acad. Sci. USA 2017, 114, 3539–3544. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Terzaghi, W.; Gong, Y.; Li, C.; Ling, J.J.; Fan, Y.; Qin, N.; Gong, X.; Zhu, D.; Deng, X.W. Modulation of BIN2 kinase activity by HY5 controls hypocotyl elongation in the light. Nat. Commun. 2020, 11, 1592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gampala, S.S.; Kim, T.W.; He, J.X.; Tang, W.; Deng, Z.; Bai, M.Y.; Guan, S.; Lalonde, S.; Sun, Y.; Gendron, J.M.; et al. An essential role for 14-3-3 proteins in brassinosteroid signal transduction in Arabidopsis. Dev. Cell 2007, 13, 177–189. [Google Scholar] [CrossRef] [Green Version]
- Hoecker, U. The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling. Curr. Opin. Plant Biol. 2017, 37, 63–69. [Google Scholar] [CrossRef]
- Liu, B.; Zuo, Z.; Liu, H.; Liu, X.; Lin, C. Arabidopsis cryptochrome 1 interacts with SPA1 to suppress COP1 activity in response to blue light. Genes Dev. 2011, 25, 1029–1034. [Google Scholar] [CrossRef] [Green Version]
- Lian, H.L.; He, S.B.; Zhang, Y.C.; Zhu, D.M.; Zhang, J.Y.; Jia, K.P.; Sun, S.X.; Li, L.; Yang, H.Q. Blue-light-dependent interaction of cryptochrome 1 with SPA1 defines a dynamic signaling mechanism. Genes Dev. 2011, 25, 1023–1028. [Google Scholar] [CrossRef] [Green Version]
- Pham, V.N.; Kathare, P.K.; Huq, E. Phytochromes and phytochrome interacting factors. Plant Physiol. 2018, 176, 1025–1038. [Google Scholar] [CrossRef] [Green Version]
- Gangappa, S.N.; Botto, J.F. The multifaceted roles of HY5 in plant growth and development. Mol. Plant 2016, 9, 1353–1365. [Google Scholar] [CrossRef] [Green Version]
- Osterlund, M.T.; Hardtke, C.S.; Wei, N.; Deng, X.W. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 2000, 405, 462–466. [Google Scholar] [CrossRef] [PubMed]
- Truernit, E.; Bauby, H.; Belcram, K.; Barthelemy, J.; Palauqui, J.C. OCTOPUS, a polarly localised membrane-associated protein, regulates phloem differentiation entry in Arabidopsis thaliana. Development 2012, 139, 1306–1315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anne, P.; Azzopardi, M.; Gissot, L.; Beaubiat, S.; Hematy, K.; Palauqui, J.C. OCTOPUS negatively regulates BIN2 to control phloem differentiation in Arabidopsis thaliana. Curr. Biol. 2015, 25, 2584–2590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pillitteri, L.J.; Peterson, K.M.; Horst, R.J.; Torii, K.U. Molecular profiling of stomatal meristemoids reveals new component of asymmetric cell division and commonalities among stem cell populations in Arabidopsis. Plant Cell 2011, 23, 3260–3275. [Google Scholar] [CrossRef] [Green Version]
- Houbaert, A.; Zhang, C.; Tiwari, M.; Wang, K.; de Marcos Serrano, A.; Savatin, D.V.; Urs, M.J.; Zhiponova, M.K.; Gudesblat, G.E.; Vanhoutte, I.; et al. POLAR-guided signalling complex assembly and localization drive asymmetric cell division. Nature 2018, 563, 574–578. [Google Scholar] [CrossRef]
- Liu, J.; Chen, J.; Zheng, X.; Wu, F.; Lin, Q.; Heng, Y.; Tian, P.; Cheng, Z.; Yu, X.; Zhou, K.; et al. GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants 2017, 3, 17043. [Google Scholar] [CrossRef]
- Hirano, K.; Kawamura, M.; Araki-Nakamura, S.; Fujimoto, H.; Ohmae-Shinohara, K.; Yamaguchi, M.; Fujii, A.; Sasaki, H.; Kasuga, S.; Sazuka, T. Sorghum DW1 positively regulates brassinosteroid signaling by inhibiting the nuclear localization of BRASSINOSTEROID INSENSITIVE 2. Sci. Rep. 2017, 7, 126. [Google Scholar] [CrossRef]
- Samakovli, D.; Margaritopoulou, T.; Prassinos, C.; Milioni, D.; Hatzopoulos, P. Brassinosteroid nuclear signaling recruits HSP90 activity. New Phytol. 2014, 203, 743–757. [Google Scholar] [CrossRef]
- Peng, P.; Yan, Z.; Zhu, Y.; Li, J. Regulation of the Arabidopsis GSK3-like kinase BRASSINOSTEROID-INSENSITIVE 2 through proteasome-mediated protein degradation. Mol. Plant 2008, 1, 338–346. [Google Scholar] [CrossRef] [Green Version]
- Lama, S.; Broda, M.; Abbas, Z.; Vaneechoutte, D.; Belt, K.; Sall, T.; Vandepoele, K.; van Aken, O. Neofunctionalization of mitochondrial proteins and incorporation into signaling networks in plants. Mol. Biol. Evol. 2019, 36, 974–989. [Google Scholar] [CrossRef]
- Failor, K.L.; Desyatnikov, Y.; Finger, L.A.; Firestone, G.L. Glucocorticoid-induced degradation of glycogen synthase kinase-3 protein is triggered by serum- and glucocorticoid-induced protein kinase and Akt signaling and controls beta-catenin dynamics and tight junction formation in mammary epithelial tumor cells. Mol. Endocrinol. 2007, 21, 2403–2415. [Google Scholar] [CrossRef] [PubMed] [Green Version]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mao, J.; Li, J. Regulation of Three Key Kinases of Brassinosteroid Signaling Pathway. Int. J. Mol. Sci. 2020, 21, 4340. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21124340
Mao J, Li J. Regulation of Three Key Kinases of Brassinosteroid Signaling Pathway. International Journal of Molecular Sciences. 2020; 21(12):4340. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21124340
Chicago/Turabian StyleMao, Juan, and Jianming Li. 2020. "Regulation of Three Key Kinases of Brassinosteroid Signaling Pathway" International Journal of Molecular Sciences 21, no. 12: 4340. https://0-doi-org.brum.beds.ac.uk/10.3390/ijms21124340




